Abstract
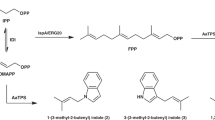
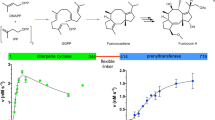
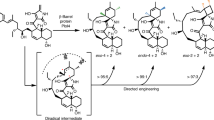
Class II terpene cyclases, such as oxidosqualene and squalene-hopene cyclases, catalyse some of the most complex polycyclization reactions. They minimally exhibit a β,γ-didomain architecture that has been evolutionarily repurposed in a wide range of terpene-processing enzymes and likely resulted from a fusion of unidentified monodomain proteins. Although single domain class I terpene cyclases have already been identified, the corresponding class II counterparts have not been previously reported. Here we present high-resolution X-ray structures of a monodomain class II cyclase, merosterolic acid synthase (MstE). With a minimalistic β-domain architecture, this cyanobacterial enzyme is able to construct four rings in cytotoxic meroterpenoids with a sterol-like topology. The structures with bound substrate, product, and inhibitor provide detailed snapshots of a cyclization mechanism largely governed by residues located in a noncanonical enzyme region. Our results complement the few known class II cyclase crystal structures, while also indicating that archaic monodomain cyclases might have already catalyzed complex reaction cascades.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Dickschat, J. S. Isoprenoids in three-dimensional space: the stereochemistry of terpene biosynthesis. Nat. Prod. Rep. 28, 1917–1936 (2011).
Rudolf, J. D. & Chang, C.-Y. Terpene synthases in disguise: enzymology, structure, and opportunities of non-canonical terpene synthases. Nat. Prod. Rep. 37, 425–463 (2020).
Wendt, K. U. & Schulz, G. E. Isoprenoid biosynthesis: manifold chemistry catalyzed by similar enzymes. Structure 6, 127–133 (1998).
Lesburg, C. A., Zhai, G., Cane, D. E. & Christianson, D. W. Crystal structure of pentalenene synthase: mechanistic insights on terpenoid cyclization reactions in biology. Science 277, 1820–1824 (1997).
Wendt, K. U., Schulz, G. E., Corey, E. J. & Liu, D. R. Enzyme mechanisms for polycyclic triterpene formation. Angew. Chem. Int. Ed. 39, 2812–2833 (2000).
Hammer, S. C., Marjanovic, A., Dominicus, J. M., Nestl, B. M. & Hauer, B. Squalene hopene cyclases are protonases for stereoselective Brønsted acid catalysis. Nat. Chem. Biol. 11, 121–126 (2015).
Cao, R. et al. Diterpene cyclases and the nature of the isoprene fold. Proteins 78, 2417–2432 (2010).
Christianson, D. W. Structural and chemical biology of terpenoid cyclases. Chem. Rev. 117, 11570–11648 (2017).
Oldfield, E. & Lin, F.-Y. Terpene biosynthesis: modularity rules. Angew. Chem. Int. Ed. 51, 1124–1137 (2012).
Köksal, M., Chou, W. K. W., Cane, D. E. & Christianson, D. W. Structure of 2-methylisoborneol synthase from Streptomyces coelicolor and implications for the cyclization of a noncanonical C-methylated monoterpenoid substrate. Biochemistry 51, 3011–3020 (2012).
Baer, P. et al. Induced-fit mechanism in class I terpene cyclases. Angew. Chem. Int. Ed. 53, 7652–7656 (2014).
Rynkiewicz, M. J., Cane, D. E. & Christianson, D. W. Structure of trichodiene synthase from Fusarium sporotrichioides provides mechanistic inferences on the terpene cyclization cascade. Proc. Natl Acad. Sci. USA 98, 13543–13548 (2001).
Janke, R., Görner, C., Hirte, M., Brück, T. & Loll, B. The first structure of a bacterial diterpene cyclase: CotB2. Acta Crystallogr. D 70, 1528–1537 (2014).
Starks, C. M., Back, K., Chappell, J. & Noel, J. P. Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science 277, 1815–1820 (1997).
Hyatt, D. C. et al. Structure of limonene synthase, a simple model for terpenoid cyclase catalysis. Proc. Natl Acad. Sci. USA 104, 5360–5365 (2007).
Köksal, M., Jin, Y., Coates, R. M., Croteau, R. & Christianson, D. W. Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis. Nature 469, 116–120 (2011).
Wendt, K. U., Poralla, K. & Schulz, G. E. Structure and function of a squalene cyclase. Science 277, 1811–1815 (1997).
Thoma, R. et al. Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase. Nature 432, 118–122 (2004).
Rudolf, J. D. et al. Structure of the ent-copalyl diphosphate synthase PtmT2 from Streptomyces platensis CB00739, a bacterial type II diterpene synthase. J. Am. Chem. Soc. 138, 10905–10915 (2016).
Köksal, M., Hu, H., Coates, R. M., Peters, R. J. & Christianson, D. W. Structure and mechanism of the diterpene cyclase ent-copalyl diphosphate synthase. Nat. Chem. Biol. 7, 431–433 (2011).
Moosmann, P. et al. Cyanobacterial ent-sterol-like natural products from a deviated ubiquinone pathway. Angew. Chem. Int. Ed. 56, 4987–4990 (2017).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Lenhart, A. et al. Binding structure and potencies of oxidosqualene cyclase inhibitors with the homologous squalene-hopene cyclase. J. Med. Chem. 46, 2083–2092 (2003).
Wendt, K. U., Lenhart, A. & Schulz, G. E. The structure of the membrane protein squalene-hopene cyclase at 2.0 Å resolution. J. Mol. Biol. 286, 175–187 (1999).
Poralla, K. et al. A specific amino acid repeat in squalene and oxidosqualene cyclases. Trends Biochem. Sci. 19, 157–158 (1994).
Corey, E. J. et al. Methodology for the preparation of pure recombinant S. cerevisiae lanosterol synthase using a baculovirus expression system. Evidence that oxirane cleavage and A-ring formation are concerted in the biosynthesis of lanosterol from 2,3-oxidosqualene. J. Am. Chem. Soc. 119, 1277–1288 (1997).
Smentek, L. & Hess, B. A. Jr. Compelling computational evidence for the concerted cyclization of the ABC rings of hopene from protonated squalene. J. Am. Chem. Soc. 132, 17111–17117 (2010).
Morikubo, N. et al. Cation–π interaction in the polyolefin cyclization cascade uncovered by incorporating unnatural amino acids into the catalytic sites of squalene cyclase. J. Am. Chem. Soc. 128, 13184–13194 (2006).
Thimmappa, R., Geisler, K., Louveau, T., O’Maille, P. & Osbourn, A. Triterpene biosynthesis in plants. Annu. Rev. Plant Biol. 65, 225–257 (2014).
Zhang, H., Khalil, Z. G. & Capon, R. J. Fascioquinols A–F: bioactive meroterpenes from a deep-water southern Australian marine sponge, Fasciospongia sp. Tetrahedron 67, 2591–2595 (2011).
Brocks, J. J., Buick, R., Summons, R. E. & Logan, G. A. A reconstruction of Archean biological diversity based on molecular fossils from the 2.78 to 2.45 billion-year-old Mount Bruce Supergroup, Hamersley Basin, Western Australia. Geochim. Cosmochim. Acta 67, 4321–4335 (2003).
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Langer, G., Cohen, S. X., Lamzin, V. S. & Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 (2008).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Holm, L. & Laakso, L. M. Dali server update. Nucleic Acids Res. 44, W351–W355 (2016).
Kraulis, P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
The PyMOL Molecular Graphics System, v.2.0 (Schrödinger, LLC, 2017).
Lang, M. & Steglich, W. An effective method for the synthesis of 13C-labeled polyprenylhydroxybenzoic acids. Synthesis 2005, 1019–1027 (2005).
Madeira, F. et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641 (2019).
Whelan, S. & Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699 (2001).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Acknowledgements
We thank R. Ueoka for support with NMR and the staff of beamline X06SA at the Paul Scherrer Institute for assistance during data collection. We acknowledge funding by the Swiss National Science Foundation (contract numbers 205321-165695 and 205320-185077) and by the Helmut Horten Foundation (to J.P.).
Author information
Authors and Affiliations
Contributions
P.M., F.E., M.G., and J.P. designed, conducted, and analysed the experiments. S.L.-M. and C.L.D. conducted organic syntheses. J.K.B.C. performed phylogenetic analyses. J.P., M.G, F.E., and P.M. wrote the paper. All authors edited the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Protein alignment.
Structure-guided alignment of merosterolic acid synthase (MstE) with squalene-hopene cyclase (SHC; PDB 2SQC; 15.91 % identity*) and oxidosqualene cyclase (OSC; PDB 1W6K; 15.26 % identity*). Alpha helices are indicated with cylinders under the sequences, beta strands with arrows. These secondary structural elements are colored yellow if they are part of the core scaffold of the γ domain and blue if they are part of β domain. Non-conserved secondary structures are depicted in grey. The catalytic aspartate residue is indicated with a red asterisk. *Identity percentages were calculated without the γ-domains of SHC and OSC.
Extended Data Fig. 2 Phylogenetic tree of selected terpene cyclase β domains.
Maximum-likelihood tree of β domains from functionally characterized and unknown terpene cyclases. Organisms are colored according to the phylogeny of their origin: Protists grey, animals gold, fungi red, plants green, and bacteria blue. Terpene enzyme class, domain architecture, and product are indicated.
Extended Data Fig. 3 Structures of MstE in complex with different ligands.
Left: Side- and top-views (rotated by 90°) of MstE (a), MstE:FS-DHB (b), MstE_D109N:GG-DHB (c), MstE_D109A:MA (d), MstE_R337A:GG-DHB (e) and MstE_Y157F (f) as cartoons with helices and β-sheets colored in pink and blue, respectively. Dashed loops indicate disordered regions in the crystal structures and are labeled accordingly. Carbon atoms of ligands, if present, are shown as sticks in gold, with the DHB moieties in black. Double bonds and newly formed C-C bonds are highlighted in black. Right: Close-up views of the active sites. The 2FO-FC electron density maps (gray mesh, contoured to 1.0 σ) of ligands are shown together with residues engaged in binding. Side-views of all compounds (rotated by 90°) are depicted on the far right. Dashed lines represent important interactions. Mutated residues are colored green. Diffuse electron density in F indicates the presence of a ligand at low occupancy.
Extended Data Fig. 4 A proton relay conveys protonation of GG-DHB.
(a) The structure of MstE:FS-DHB superpositioned with MstE_D109N:GG-DHB (RMSD-Cα: 0.12 Å) reveals that Asp109 adopts anti-position for protonation of the terminal C = C in GG-DHB. Two water molecules align between Tyr216 and Asp109 to activate the Brønsted acid and replace the proton after reaction. Oxygen atoms are highlighted in red; the protons (light blue) were added with PyMol (37) and oriented according to potential interactions represented as dashed lines (distance given in Å). (b) The primary amine of Asn109 in MstE_D109N:GG-DHB displaces one of the water molecules by 0.6 Å, flipping the orientation of all protons in the cluster.
Extended Data Fig. 5 A charge relay supports deprotonation of GG-DHB.
The structure of MstE_D109N:GG-DHB reveals the formation of a triad between Glu339, Arg337 and Asp162. Asp162 forms a salt bridge with the guanidino group of Arg337. This pushes electrons towards the secondary amine which in turn polarizes the carboxylate of Glu339, strengthening the H-bond with the meta-hydroxy group of DHB and increasing basicity. After deprotonation, this cluster acts as a proton-relay, before it is promptly broken and reordered to accommodate the product. Heteroatoms are highlighted in red (O) and blue (N); the proton (light blue) was added with PyMol. Interactions are represented as dashed lines (distance given in Å).
Supplementary information
Supplementary Information
Supplementary Methods, Supplementary Figs. 1–36, and Supplementary Tables 1–4.
Rights and permissions
About this article
Cite this article
Moosmann, P., Ecker, F., Leopold-Messer, S. et al. A monodomain class II terpene cyclase assembles complex isoprenoid scaffolds. Nat. Chem. 12, 968–972 (2020). https://doi.org/10.1038/s41557-020-0515-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-020-0515-3
This article is cited by
-
Structure-guided product determination of the bacterial type II diterpene synthase Tpn2
Communications Chemistry (2022)