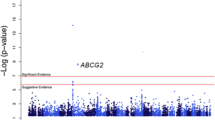
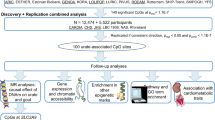
Abstract
Gout is a complex inflammatory arthritis affecting ~20% of people with an elevated serum urate level (hyperuricemia). Gout and hyperuricemia are essentially specific to humans and other higher primates, with varied prevalence across ancestral groups. SLC2A9 and ABCG2 are major loci associated with both urate and gout in multiple ancestral groups. However, fine mapping has been challenging due to extensive linkage disequilibrium underlying the associated regions. We used trans-ancestral fine mapping integrated with primate-specific genomic information to address this challenge. Trans-ancestral meta-analyses of GWAS cohorts of either European (EUR) or East Asian (EAS) ancestry resulted in single-variant resolution mappings for SLC2A9 (rs3775948 for urate and rs4697701 for gout) and ABCG2 (rs2622621 for gout). Tests of colocalization of variants in both urate and gout suggested existence of a shared candidate causal variant for SLC2A9 only in EUR and for ABCG2 only in EAS. The fine-mapped gout variant rs4697701 was within an ancient enhancer, whereas rs2622621 was within a primate-specific transposable element, both supported by functional evidence from the Roadmap Epigenomics project in human primary tissues relevant to urate and gout. Additional primate-specific elements were found near both loci and those adjacent to SLC2A9 overlapped with known statistical epistatic interactions associated with urate as well as multiple super-enhancers identified in urate-relevant tissues. We conclude that by leveraging ancestral differences trans-ancestral fine mapping has identified ancestral and functional variants for SLC2A9 or ABCG2 with primate-specific regulatory effects on urate and gout.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Change history
08 September 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Dalbeth N, Merriman TR, Stamp LK. Gout. Lancet. 2016;388:2039–52.
Campion EW, Glynn RJ, Delabry LO. Asymptomatic hyperuricemia. Risks and consequences in the normative aging study. Am J Med. 1987;82:421–6.
Dalbeth N, Phipps-Green A, Frampton C, Neogi T, Taylor WJ, Merriman TR. Relationship between serum urate concentration and clinically evident incident gout: an individual participant data analysis. Ann Rheum Dis. 2018;77:1048–52.
Kuo CF, Grainge MJ, Zhang W, Doherty M. Global epidemiology of gout: prevalence, incidence and risk factors. Nat Rev Rheumatol. 2015;11:649–62.
Köttgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet. 2013;45:145–54.
Cadzow M, Merriman TR, Dalbeth N. Performance of gout definitions for genetic epidemiological studies: Analysis of UK Biobank. Arthritis Res Ther. 2017;19:181.
Kanai M, Akiyama M, Takahashi A, Matoba N, Momozawa Y, Ikeda M, et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat Genet. 2018;50:390–400.
Nakatochi M, Kanai M, Nakayama A, Hishida A, Kawamura Y, Ichihara S, et al. Genome-wide meta-analysis identifies multiple novel loci associated with serum uric acid levels in Japanese individuals. Commun Biol. 2019;2:115.
Tin A, Marten J, Halperin Kuhns VL, Li Y, Wuttke M, Kirsten H, et al. Target genes, variants, tissues and transcriptional pathways influencing human serum urate levels. Nat Genet. 2019;51:1459–74.
Major TJ, Dalbeth N, Stahl EA, Merriman TR. An update on the genetics of hyperuricaemia and gout. Nat. Rev. Rheumatol. 2018;14:351–3.
Okada Y, Sim X, Go MJ, Wu J-Y, Gu D, Takeuchi F, et al. Meta-analysis identifies multiple loci associated with kidney function-related traits in east Asian populations. Nat Genet. 2012;44:904–9.
Li C, Li Z, Liu S, Wang C, Han L, Cui L, et al. Genome-wide association analysis identifies three new risk loci for gout arthritis in Han Chinese. Nat Commun. 2015;6:7041.
Li Z, Zhou Z, Hou X, Lu D, Yuan X, Lu J, et al. Replication of Gout/Urate concentrations GWAS susceptibility loci associated with gout in a Han Chinese population. Sci Rep. 2017;7:4094.
Matsuo H, Yamamoto K, Nakaoka H, Nakayama A, Sakiyama M, Chiba T, et al. Genome-wide association study of clinically defined gout identifies multiple risk loci and its association with clinical subtypes. Ann Rheum Dis. 2016;75:652–9.
Phipps-Green AJ, Merriman ME, Topless R, Altaf S, Montgomery GW, Franklin C, et al. Twenty-eight loci that influence serum urate levels: analysis of association with gout. Ann Rheum Dis. 2016;75:124–30.
Tin A, Woodward OM, Kao WHL, Liu CT, Lu X, Nalls MA, et al. Genome-wide association study for serum urate concentrations and gout among African Americans identifies genomic risk loci and a novel URAT1 loss-of-function allele. Hum Mol Genet. 2011;20:4056–68.
Yang B, Mo Z, Wu C, Yang H, Yang X, He Y, et al. A genome-wide association study identifies common variants influencing serum uric acid concentrations in a Chinese population. BMC Med Genom. 2014;7:10.
Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373:895–907.
Gazal S, Finucane HK, Furlotte NA, Loh PR, Palamara PF, Liu X, et al. Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection. Nat Genet. 2017;49:1421–7.
Gazal S, Loh PR, Finucane HK, Ganna A, Schoech A, Sunyaev S, et al. Functional architecture of low-frequency variants highlights strength of negative selection across coding and non-coding annotations. Nat Genet. 2018;50:1600–7.
Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–43.
Woodward OM, Kottgen A, Coresh J, Boerwinkle E, Guggino WB, Kottgen M. Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proc Natl Acad Sci USA. 2009;106:10338–42.
Matsuo H, Takada T, Ichida K, Nakamura T, Nakayama A, Ikebuchi Y, et al. Common defects of ABCG2, a high-capacity urate exporter, cause gout: a function-based genetic analysis in a Japanese population. Sci Transl Med. 2009;1:5ra11.
Cleophas MC, Joosten LA, Stamp LK, Dalbeth N, Woodward OM, Merriman TR. ABCG2 polymorphisms in gout: Insights into disease susceptibility and treatment approaches. Pharmacogenom Personal Med. 2017;10:129–42.
Wrigley R, Phipps-Green AJ, Topless RK, Major TJ, Cadzow M, Riches P, et al. Pleiotropic effect of the ABCG2 gene in gout: involvement in serum urate levels and progression from hyperuricemia to gout. Arthritis Res Ther. 2020;22:45.
Friedman TB, Polanco GE, Appold JC, Mayle JE. On the loss of uricolytic activity during primate evolution-I. Silencing of urate oxidase in a hominoid ancestor. Comp Biochem Physiol—B: Comp Biochem. 1985;81:653–9.
Oda M, Satta Y, Takenaka O, Takahata N. Loss of urate oxidase activity in hominoids and its evolutionary implications. Mol Biol Evol. 2002;19:640–53.
Wu X, Wakamiya M, Vaishnav S, Geske R, Montgomery C Jr, Jones P, et al. Hyperuricemia and urate nephropathy in urate oxidase-deficient mice. Proc Natl Acad Sci USA. 1994;91:742–6.
Lu J, Dalbeth N, Yin H, Li C, Merriman TR, Wei WH. Mouse models for human hyperuricaemia: a critical review. Nat Rev Rheumatol. 2019;15:413–26.
Doan RN, Bae BI, Cubelos B, Chang C, Hossain AA, Al-Saad S, et al. Mutations in human accelerated regions disrupt cognition and social behavior. Cell. 2016;167:341–54. e12.
Leask M, Dowdle A, Salvesen H, Topless R, Fadason T, Wei W, et al. Functional urate-associated genetic variants influence expression of lincRNAs LINC01229 and MAFTRR. Front Genet. 2019;9:733.
Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M. Genome-wide association studies in diverse populations. Nat Rev Genet. 2010;11:356–66.
Morris AP. Transethnic meta-analysis of genomewide association studies. Genet Epidemiol. 2011;35:809–22.
Magi R, Horikoshi M, Sofer T, Mahajan A, Kitajima H, Franceschini N, et al. Trans-ethnic meta-regression of genome-wide association studies accounting for ancestry increases power for discovery and improves fine-mapping resolution. Hum Mol Genet. 2017;26:3639–50.
Fortune MD, Guo H, Burren O, Schofield E, Walker NM, Ban M, et al. Statistical colocalization of genetic risk variants for related autoimmune diseases in the context of common controls. Nat Genet. 2015;47:839–46.
Rasmussen MD, Hubisz MJ, Gronau I, Siepel A. Genome-wide inference of ancestral recombination graphs. PLoS Genet. 2014;10:e1004342.
Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7.
Backenroth D, He ZH, Kiryluk K, Boeva V, Pethukova L, Khurana E, et al. FUN-LDA: a latent dirichlet allocation model for predicting tissue-specific functional effects of noncoding variation: methods and applications. Am J Hum Genet. 2018;102:920–42.
Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–94.
Emera D, Yin J, Reilly SK, Gockley J, Noonan JP. Origin and evolution of developmental enhancers in the mammalian neocortex. Proc Natl Acad Sci USA. 2016;113:E2617–26.
Kronenberg ZN, Fiddes IT, Gordon D, Murali S, Cantsilieris S, Meyerson OS, et al. High-resolution comparative analysis of great ape genomes. Science. 2018;360:eaar6343.
Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98.
Ma W, Ay F, Lee C, Gulsoy G, Deng X, Cook S, et al. Fine-scale chromatin interaction maps reveal the cis-regulatory landscape of human lincRNA genes. Nat Methods. 2015;12:71–8.
Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–40.
Trizzino M, Park Y, Holsbach-Beltrame M, Aracena K, Mika K, Caliskan M, et al. Transposable elements are the primary source of novelty in primate gene regulation. Genome Res. 2017;27:1623–33.
Wallace SL, Robinson H, Masi AT, Decker JL, McCarty DJ, Yu TF. Preliminary criteria for the classification of the acute arthritis of primary gout. Arthritis Rheumatol. 1977;20:895–900.
Rasheed H, McKinney C, Stamp LK, Dalbeth N, Topless RK, Day R, et al. The toll-like receptor 4 (TLR4) variant rs2149356 and risk of gout in European and polynesian sample sets. PLoS ONE. 2016;11:e0147939.
Choi HK, Curhan G. Coffee consumption and risk of incident gout in women: the Nurses’ Health Study. Am J Clin Nutr. 2010;92:922–7.
McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–83.
Loh PR, Danecek P, Palamara PF, Fuchsberger C, Y AR, H KF, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet. 2016;48:1443–8.
Durbin R. Efficient haplotype matching and storage using the positional Burrows-Wheeler transform (PBWT). Bioinformatics. 2014;30:1266–72.
Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499.
Magi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinform. 2010;11:288.
Horikoshi M, Pasquali L, Wiltshire S, Huyghe JR, Mahajan A, Asimit JL, et al. Transancestral fine-mapping of four type 2 diabetes susceptibility loci highlights potential causal regulatory mechanisms. Hum Mol Genet. 2016;25:2070–81.
Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, Ferreira T, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–44.
Wellcome Trust Case Control C, Maller JB, McVean G, Byrnes J, Vukcevic D, Palin K, et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44:1294–301.
Popadin Konstantin Y, Gutierrez-Arcelus M, Lappalainen T, Buil A, Steinberg J, Nikolaev Sergey I, et al. Gene age predicts the strength of purifying selection acting on gene expression variation in humans. Am J Hum Genet. 2014;95:660–74.
Zhang YE, Vibranovski MD, Landback P, Marais GA, Long M. Chromosomal redistribution of male-biased genes in mammalian evolution with two bursts of gene gain on the X chromosome. PLoS Biol. 2010;8:e1000494.
Yang DC, Jang I, Choi J, Kim MS, Lee AJ, Kim H, et al. 3DIV: A 3D-genome interaction viewer and database. Nucleic Acids Res. 2018;46(D1):D52–7.
Wei W-H, Guo Y, Kindt AS, Merriman TR, Semple CA, Wang K, et al. Abundant local interactions in the 4p16.1 region suggest functional mechanisms underlying SLC2A9 associations with human serum uric acid. Hum Mol Genet. 2014;23:5061–8.
Tin A, Li Y, Brody JA, Nutile T, Chu AY, Huffman JE, et al. Large-scale whole-exome sequencing association studies identify rare functional variants influencing serum urate levels. Nat Commun. 2018;9:4228.
Dao LTM, Galindo-Albarrán AO, Castro-Mondragon JA, Andrieu-Soler C, Medina-Rivera A, Souaid C, et al. Genome-wide characterization of mammalian promoters with distal enhancer functions. Nat Genet. 2017;49:1073.
Brodie A, Azaria JR, Ofran Y. How far from the SNP may the causative genes be? Nucleic Acids Res. 2016;44:6046–54.
Vargas-Santos AB, Taylor WJ, Neogi T. Gout classification criteria: update and implications. Curr Rheumatol Rep. 2016;18:46.
Prescott SL, Srinivasan R, Marchetto MC, Grishina I, Narvaiza I, Selleri L, et al. Enhancer divergence and cis-regulatory evolution in the human and chimp neural crest. Cell. 2015;163:68–83.
Acknowledgements
This research has been conducted using the UK Biobank Resource under Application Number 12611. RT is funded by the University of Otago Research Grant awarded to TRM and WHW. Health Research Council NZ funded LKS, ND, TRM and WHW’s time in part (HRC 19/206). WHW is funded by Cure Kids NZ and the University of Otago and Health Research Council NZ (HRC 17/288). WHW is very grateful for the important support provided by Professor Stephen Robertson and the Clinical Genetics Group in the University of Otago, Dunedin, New Zealand.
Eurogout consortium
A. Abhishek, M. Andres, T. Crisan, N. Dalbeth, M. Doherty, L. Jacobsson, M. Janssen, T.L. Jansen, L.A. Joosten, M. Kapetanovic, F. Lioté, H. Matsuo, G. McCarthy, T. Merriman, F. Perez-Ruiz, P.L. Riches, P. Richette, P.C. Robinson, E. Roddy, B. Stiburkova, A. So, L.K. Stamp, A.K. Tausche, R. Torres-Jiminez, T. Uhlig.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Conflict of interest
ND disclosures: ND has received consulting fees, speaker fees or grants from AstraZeneca, Horizon, Amgen, Selecta, Arthrosi, Dyve BioSciences, Hengrui. Abbvie, and Janssen, outside the submitted work. The other authors declare no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Members of the Eurogout Consortium are listed below Acknowledgements.
Supplementary information
Rights and permissions
About this article
Cite this article
Takei, R., Cadzow, M., Markie, D. et al. Trans-ancestral dissection of urate- and gout-associated major loci SLC2A9 and ABCG2 reveals primate-specific regulatory effects. J Hum Genet 66, 161–169 (2021). https://doi.org/10.1038/s10038-020-0821-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-020-0821-z
This article is cited by
-
Global status and trends in gout research from 2012 to 2021: a bibliometric and visual analysis
Clinical Rheumatology (2023)