Abstract
The accumulation of abnormal prion protein (PrPSc) produced by the structure conversion of PrP (PrPC) in the brain induces prion disease. Although the conversion process of the protein is still not fully elucidated, it has been known that the intramolecular chemical bridging in the most fragile pocket of PrP, known as the “hot spot,” stabilizes the structure of PrPC and inhibits the conversion process. Using our original structure-based drug discovery algorithm, we identified the low molecular weight compounds that predicted binding to the hot spot. NPR-130 and NPR-162 strongly bound to recombinant PrP in vitro, and fragment molecular orbital (FMO) analysis indicated that the high affinity of those candidates to the PrP is largely dependent on nonpolar interactions, such as van der Waals interactions. Those NPRs showed not only significant reduction of the PrPSc levels but also remarkable decrease of the number of aggresomes in persistently prion-infected cells. Intriguingly, treatment with those candidate compounds significantly prolonged the survival period of prion-infected mice and suppressed prion disease-specific pathological damage, such as vacuole degeneration, PrPSc accumulation, microgliosis, and astrogliosis in the brain, suggesting their possible clinical use. Our results indicate that in silico drug discovery using NUDE/DEGIMA may be widely useful to identify candidate compounds that effectively stabilize the protein.
Similar content being viewed by others
Introduction
Prion disease, also known as transmissible spongiform encephalopathy, is a zoonotic infectious disease that affects both humans and animals, such as bovine, deer, goats, and sheep. The pathogenic mechanism of this disease involves the conversion of a normal prion protein (PrPC) within an organism to an abnormal prion protein (PrPSc). Symptoms are caused by the accumulation of PrPSc as an insoluble amyloid in the brain. Compared with PrPC, PrPSc forms a stable amyloid structure characterized by an extensive rich β-sheet secondary structural component and has characteristic features, such as partial resistance to proteinase K digestion. Furthermore, unlike typical pathogens, such as bacteria and viruses, the prion pathogen cannot be inactivated by UV and X-ray because it is composed of protein only (without nucleic acid).
Sporadic Creutzfeldt–Jakob disease (sCJD) makes up ~ 80% of human prion diseases, and the cause remains unknown. The pathological condition begins with dementia and ends with death after a persistent vegetative state. In the pathological processes, there are the cases are that are rapidly progressing (< 1 month) and those that can persist over several years. CJD is a type of rare lethal neurodegenerative disorder, but it can affect everyone. In a rapidly progressing illness state, CJD patients place a heavy burden on family and healthcare workers. Moreover, handling CJD patients is challenging for healthcare workers because the blood and tissues of these patients may be infected. Prion drugs, which enable PrPSc reduction by acting on a variety of different mechanisms, have been reported and described as those that downregulate PrPC by change of PrPC localization in the cell by pentosan polysulfate (PPS) [1], amphotericin B [2], filipin [3], and suramin [4]; promote PrPC degradation by IU-1 [5]; stabilize PrPC conformation by GN8 [6], MS [7], and NPRs [8]; inhibit the interaction between PrPC and PrPSc by anti-PrP antibodies, such as Fab D18 [9], 6H4 [10], and ICSM18 [11]; inhibit PrPSc polymerization and accumulation by beta-sheet breaker iPrP13 [12]; destabilize PrPSc by polypropylene imine [13], tetracycline, and doxycycline (DOXY) [14, 15]; inhibit structural conversion from PrPC to PrPSc by peptide as PrP106–128, PrP113–141 [16], and Congo Red [17]; and enhance PrPSc degradation by lithium [18], trehalose [19], rapamycin [20], FK506 [21], and tamoxifen [22]. Among them, PSS, amphotericin B, and DOXY used to treat prion-infected animals exhibited a clear therapeutic effect against progressive clinical signs of neurological illness [15, 23, 24]; however, no therapeutic effect was observed following treatment of CJD patients with those drugs [25,26,27]. Thus far, the therapeutic agent for prion disease has not been developed.
Novel drug discovery is performed with structure-based drug design (SBDD) approaches. SBDD is a concept in which the discovered candidate compounds bind to the target proteins of the diseases by virtual screening with computer simulation based on protein structure information, and it rapidly achieves results in identifying effective compounds to treat several diseases, such as raltitrexed, which targets thymidylate synthase [28], and amprenavir, which targets antiretroviral protease [29, 30], for human immunodeficiency virus; isoniazid, which targets InhA for tuberculosis [31]; flurbiprofen, which targets cyclooxygenase-2 for rheumatoid arthritis and osteoarthritis [32, 33]; STX-0119, which targets STAT3 for lymphoma [34]; and dorzolamide, which targets carbonic anhydrase for glaucoma and cystoid macular edema [35].
Recently, novel compounds for treating prion disease were reported: 1) Cp-60 inhibited PrPSc replication by acting as dominant-negative on PrPC [36] and 2) GN8 and the GN8-optimized derivative (MC) inhibited structure conversion from PrPC to PrPSc by strongly stabilizing the conformation after binding to PrPC and showed medicinal effect for prion-infected animals [6, 7]. In addition, BMDs that strongly interact with the binding site of GN8 in PrPC were identified by virtual screening with the original simulation approach, and the compounds suppressed PrPSc production in prion-infected cells [37]. We also previously reported that NPRs identified by the SBDD approach with original docking simulation software, called Nagasaki University Docking Engine (NUDE), have potential as novel therapeutic candidates for prion disease [8]. Our virtual screening can calculate the binding affinities between not only PrPC but also other targeting proteins for several diseases and small compounds in a large chemical compound database using the DEstination for Gpu Intensive MAchine (DEGIMA) supercomputer system [38]. The NPRs significantly reduced PrPSc levels in prion-infected cells in a dose-dependent manner, and their IC50 equaled that of GN8. NPRs reduced PrPSc levels in prion-infected mice brain at 110 days post infection (d.p.i.) before onset, but did not prolong survival periods of the prion-infected mice [8].
In this study, to enhance the accuracy of our docking simulation, we conducted an analysis by increasing the number of compounds and limiting the binding region of the compound to PrPC. Following virtual screening, binding interactions of candidate compounds to PrPC were evaluated using surface plasmon resonance (SPR) analysis. Consecutively, to investigate whether candidate compounds have potential as therapeutic agents for prion disease, we analyzed the anti-prion effect of the compounds using prion-infected in vitro and in vivo models.
Materials and Methods
Regents and Antibodies
All compounds (#01 to #73) were purchased from ASINEX (NC, USA). The compounds were completely dissolved in dimethyl sulfoxide (DMSO). To detect proteinase K-digested PrPSc in prion-infected cells and mice brains and spleens, M20 (Santa Cruz Biotechnology, CA, USA) and SAF83 (SPI bio, Montigny Le Bretonneux, France) were used for immunoblotting. β-Actin (MBL, Nagoya, Japan) was used for loading control in immunoblotting. SAF61 (SPI bio), which detects denatured PrPSc, was used for immunofluorescence staining. SAF32 (SPI bio) detecting PrPSc, Iba-1 (WAKO, Osaka, Japan) detecting microglia, and glial fibrillary acidic protein (GFAP) (DAKO, Glostrup, Denmark) detecting astrocyte were used for immunohistochemistry. Those antibodies were purchased from the indicated vendors. As secondary antibodies, goat anti-horseradish peroxidase conjugates (Jackson ImmunoResearch, Cambridge, UK) were used for immunoblotting. EnVision polymer horseradish peroxidase-conjugated anti-rabbit and anti-mouse IgG antibodies (Dako, Rabbit: K4002, Mouse: K4000) were used for immunohistochemistry.
In Silico Screening
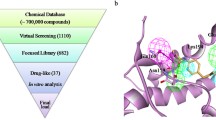
To determine candidate anti-prion compounds, we performed a docking simulation using an original chemical compound library that included ~ 690,000 compounds. In the docking simulation, a structure information of human PrPC (125th–230th amino acid residues) was used, and the information was provided by nuclear magnetic resonance spectroscopy [39] (protein data band code: 2LSB). To calculate the binding affinity between PrPC and compounds, the search region of the docking simulation was limited to a small cubic space (15 Å × 15 Å × 15 Å), which included the PrPC “hot spot” that selectively binds to the anti-prion compound GN8 [6, 40]. The calculation was performed using the DEGIMA supercomputer’s graphics processing unit (GPU)-designed docking simulation program. The DEGIMA supercomputer was constructed with more than 100 GPUs, as previously described [8, 38].
Fragment Molecular Orbital Calculation
To analyze the binding structure obtained from the docking simulation, ab initio quantum chemical calculations based on the fragment molecular orbital (FMO) method [41] were performed. To prepare appropriate atomic coordinates for the FMO calculations, hydrogen atoms were added to the docking structure, and the N-terminal (LEU125) and C-terminal (SER230) were capped by –COCH3 and –NHCH3, respectively, which was followed by a classical energy minimization using FF14SB [42] and GAFF [43] force fields. In the FMO calculations, the amino acid residues and the compound were treated as a single fragment except for cysteines forming a disulfide bond, which were merged into one fragment. Interaction energies were calculated at the Hartree–Fock (HF) and second-order Møller–Plesset perturbation (MP2) levels, with the resolution of the identity approximation [44] using the cc-pVDZ basis set [45]. In this study, PAICS [46] was used for the FMO calculations.
SPR Analysis
The binding affinity between recombinant PrP and compounds were analyzed using a Biacore T200 system (GE Healthcare, MA, USA) as previously described [8]. The procedure of recombinant human and mouse PrP (23rd to 231st amino acid residues) synthesis was carried out as previously described [47]. Briefly, the recombinants were synthesized by Escherichia coli (BL21) using the prnp-coded pET plasmids and purified by imidazole. Those recombinants acting as ligands were immobilized on a CM5 sensor chip (GE, BR-100530) using an Amine Coupling Kit (GE, BR-1000-50), according to the manufacturer’s protocol. Individual compounds were adjusted at 10 μM with a running buffer (10 mM HEPES, pH 7.4 containing 150 mM NaCl, 0.05% Tween 20, and 5% DMSO), and we injected them sequentially for 2 min at a flow rate of 30 mL/min. Data were corrected by subtracting the value of a blank sensor chip as a background from individual measured values.
Prion-Infected Cell Culture Model
Persistently infected prion-infected cells (N2a-FK) were established from N2a-58 cells that had been infected with a mouse-adapted Gerstmann–Sträussler–Scheinker strain (Fukuoka-1) and constitutively produced PrPSc, as previously described [20, 48]. The N2a-58 cells are established from the cells overexpressing PrPC and integrating mouse prnp gene in mouse neuroblastoma Neuro2a (N2a) cells. Those cells were cultured at 37 °C in 5% CO2 in Dulbecco’s modified Eagle’s medium containing 4500 mg/mL glucose, 10% heat-inactivated fetal bovine serum, 100 units/mL penicillin, and 100 μg/mL streptomycin. To determine the cell viability after treatment with the compounds, the proportions of live and dead cells were determined by cell staining with Trypan Blue solution.
Immunoblotting
The culture cells treated with various experimental conditions were lysed in 1× Triton-DOC lysis buffer. For PrPSc analysis, the cell lysates were digested with 20 μg/mL proteinase K, followed by the addition of SDS-sample buffer. The adjusted samples were loaded to 15% SDS-PAGE gel and subsequently blotted to a PVDF membrane. M20 and SAF83 were used as primary antibodies, and anti-goat IgG-HRP and anti-mouse IgG-HRP were used as secondary antibodies, respectively. β-Actin and anti-mouse IgG-HRP were used as primary and secondary antibodies for gel loading control. Bands were visualized using Clarity Western ECL Blotting Substrate (Bio-Rad, CA, USA), following quantification of band intensities and were measured using the ImageJ software (National Institutes of Health, MD, USA). Further details were previously described [8].
Immunofluorescence Staining
PrPSc detection in the prion-infected cells was performed as previously described [20, 49]. Cells that were cultured for 48 h after treatments were washed with PBS and fixed with 4% formaldehyde. After permeabilization using 0.5% Triton X-100, the cells were denatured with 3 M guanidine thiocyanate. They were blocked for 1 h in 5% skim milk at room temperature and incubated overnight at 4 °C with a SAF61 primary antibody (1:200), followed by incubation with an Alexa Fluor 488-conjugated anti-mouse IgG secondary antibody (Invitrogen, CA, USA) (1:500) for 1 h at 37 °C. The cells were mounted with a mounting medium containing DAPI to stain nuclei (Vector Laboratories, CA, USA). For the detection of protein aggregation, the cells were treated with a dual detection reagent ProteoStat aggresome detection kit (Enzo Life Science, NY, USA) (1:2000 ProteoStat, 1:1000 Hoechst 33342 for nuclei staining). The protein aggregation assay was performed as previously described [8]. All reagents were prepared according to the manufacturer’s instructions. All images were observed using a confocal laser-scanning microscope LSM 700 (Carl Zeiss, Oberkochen, Germany).
Prion-Infected Animal Model
In vivo prion infection animal models were prepared as previously described [8]. We adjusted NPRs with Kolliphor HS 15 (BASF, Ludwigshafen, Germany), which is one of the solvents used as pharmaceutical applications, and those compounds were dissolved in PBS including 20% Kolliphor HS 15. Four-week-old CD1 male mice were purchased from SLC (Hamamatsu, Japan) and intracerebrally inoculated with 20 μL of a 10% brain homogenate prepared from mice that had been terminally infected with the FK-1 strain. Mice were intraperitoneally administered 2.0 mg compound/kg/day every other day from shortly after prion inoculation until the terminal stage of the disease. Control mice group were treated with PBS. The mice were monitored for disease progression and sacrificed at 100 d.p.i. before clinical onset and terminal stage, and the brains and spleens were collected for histological analysis.
Pathological Analysis
Histopathological analysis was conducted as previously described [8, 50, 51]. Briefly, 3-μm-thick sections were cut from the fixed brain tissues embedded in paraffin. Spongiform changes were analyzed by staining with hematoxylin and eosin. Gliosis were evaluated by IHC staining with Iba-1 and GFAP antibodies. PrPSc was detected by IHC staining with SAF32 antibody after preparing the samples by the hydrolytic autoclaving and formic acid method [50]. As secondary antibodies, EnVision polymer horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG antibodies were used, followed by visualization with 3,3-diaminobenzidine.
Ethics Statement
Animal experiments were approved by the Animal Care and Use Committee of the Nagasaki University. All animals were treated and cared for in accordance with the Guidelines for Animal Experimentation of Nagasaki University.
Statistical Analysis
Statistical analysis of all data was performed using Statcel 2 of the Excel and GraphPad Prism software. The graph and numerical data results represent the mean ± standard deviation (SD) and standard error of the mean (SEM) of at least three independent experiments. Detailed information was previously described [51] and can be found in the figure legends.
Results
Selection of Candidate Compounds by In Silico Screening
Candidate compounds were ranked by docking scores, which show the binding affinity of these compounds with hot spot region of human PrPC from ~ 690,000 compounds. The compound library is a collection of low molecular weight (MW) compounds, which are ~ 400–600 MW. We focused on the compounds ranked in top 73 and investigated whether those compounds have potential as anti-prion agents; there was no overlap between these 73 and the top 96 from the previous screening selected from ~ 290,000 compounds [8].
Binding Ability Analysis of Candidate Compounds Against PrPC
To confirm the interactions between PrPC and the compounds deduced by virtual screening, we assessed the direct binding affinity against human and mouse PrPC using SPR analysis. The 73 compounds were adjusted to a concentration of 10 μM and sequentially underwent high-throughput screening using human or mouse PrPC solid-phase sensor chips in a Biacore apparatus. The quantification of binding affinity was evaluated as RU value, which implies binding response and strength. The ratios of compounds, which showed responses of 0 RU or less, were 20.1% (15 out of 73 compounds: for human PrP) and 28.7% (21 out of 73 compounds: for mouse PrP). On the other hand, compounds of ~ 12% (9 out of 73 compounds) showed an intensity of more than 10 RU and were confirmed to bind to recombinant human PrPC. Moreover, compounds which showed an intensity of more than 10 RU were ~ 9.6% (7 out of 73 compounds) in SPR analysis recombinant mouse PrPC. NPR-130 and NPR-162, in particular, had high responses for human and mouse PrPC, and those RU values were at least two to three times higher than others (Supplementary Fig. 1). Following a detailed kinetic analysis of NPR-130 and NPR-162 compounds with recombinant human and mouse PrPs, the sensorgram slopes of both compounds showed that the compounds dose dependently bound to the PrPs. In addition, KD values showed that the binding affinity of both compounds was stronger against mouse PrP than human PrP (Table 1). These results indicate that candidate compounds calculated by our in silico system had an ability binding to PrPC in not only virtual simulation but also SPR analysis (Fig. 1).
FMO Analysis of Molecular Interactions Between PrPC and Candidate Compounds
The interaction energies of NPR-130 and NPR-162 with 18 amino acid residues, which are located within 4.0 Å from the compounds, were calculated by the FMO method (Fig. 2). The interaction energies with a negative and positive value represent attractive and repulsive interactions, respectively, and energy obtained from the HF and MP2 levels mainly reflect a polar and nonpolar interaction, respectively. We note that the HF interaction energies (blue) of some residues were positively large, indicating that the polar interaction of both candidates is repulsive. On the other hand, interaction energies obtained by the sum of HF and MP2 energies (red) were overall negative, which suggests that the repulsive nature of the polar interaction is reversed to an attractive nonpolar interaction. Consequently, the presence of a nonpolar interaction is considered to be a major factor contributing to the high affinity of the candidates to the “hot spot” of PrPC.
Interaction energies of candidate compounds with amino acid residues. Interaction energies of NPR-130 (a) and NPR-162 (b) with the amino acid residues of PrPC located within 4.0 Å from the compounds were calculated by the FMO method. The blue value indicates the HF level interaction energy, which is mainly caused by a polar interaction. However, the red value indicates the MP2 level interaction energy, in which a nonpolar interaction is additionally considered. The binding structure of NPR-130 and NPR-162 obtained from docking simulations are also given, in which PrPC is presented in a cartoon model, and the compounds are presented in a stick model
Effect of Candidate Compounds on PrPSc in Persistently Prion-Infected Cells
To assess the effect of the compounds on PrPSc, we performed immunoblotting analysis and increased the PrPSc levels in individual lysates of persistently Gerstmann–Sträussler–Scheinker syndrome-derived Fukuoka-1 prion-infected cells (N2a-FK) that were incubated for 48 h following treatment with 10 μM compounds. The results revealed that the PrPSc levels in prion-infected cells were clearly decreased by treatment with several compounds, including NPR-130, NPR-162, and NPR-163, compared with the controls (cells using DMSO only as the solvent) (Fig. 3a and Supplementary Fig. 2). The quantification of PrPSc levels in N2a-FK cells following candidate compound treatment revealed that 17 compounds exhibited anti-prion effects, which significantly reduced PrPSc levels in vitro (compounds given in red color in Fig. 3b). In particular, 14 compounds, except NPR-111, NPR-163, and NPR-169, had strong inhibitory effects that suppressed PrPSc levels to 50% or less (Fig. 3b). These results suggested that putative compounds, which were virtually simulated using the NUDE on the DEGIMA supercomputer, had a suppressing ability on the PrPSc levels in prion-infected cells with a high probability (17 out of 73 compounds).
Anti-prion screening of candidate compounds using prion-infected cell models. (a) Effect of candidate compounds (NPR-101 to NPR-173) on PrPSc in N2a-FK cells. The cells were treated with 10 μM compounds for 48 h. PrPSc levels were validated using immunoblotting. Panels show the representative results of the immunoblots (NPR-126 to NPR-130 and NPR-160 to NPR-164). The control lane indicates lysates of DMSO-treated cells as negative controls. The lane numbers indicate the individual candidate compounds in each panel. (b) The graph shows quantifications of PrPSc band intensities in respective compound-treated N2a-FK cells for 48 h. The screening of candidate compounds was performed at least in five independent trials. Statistical significances were determined using one-way ANOVA, followed by the Tukey–Kramer test for multiple comparisons. *P < 0.05 and **P < 0.01 vs control. Data are presented as mean ± SEM
Dose Responses of Candidate Compounds Against PrPSc in Persistently Prion-Infected Cells
To evaluate the dose-dependent responses of the 17 candidate compounds, we measured PrPSc levels in cells after NPR treatment with several concentrations (0.1, 0.5, 1, 5, and 10 μM) for 48 h (Fig. 4a and Supplementary Fig. 3). PrPSc levels were dramatically decreased after 10 μM NPR-130 and NPR-162 treatments in prion-infected cells. However, those compounds did not show a clear dose–response effect (Fig. 4a). Based on these results, we determined the concentration of the 17 compounds that achieved a 50% reduction (IC50) of PrPSc, which were between 2.7 and 8.3 μM (Table 2). Following immunofluorescent staining with an anti-PrP antibody, the pretreatment of guanidine thiocyanate enables the detection of aggregated PrPSc in persistently prion-infected cells [5, 20]. A remarkable reduction in aggregated PrPSc levels was observed in the prion-infected cells after NPR-130 and NPR-162 treatments for 48 h compared with the DMSO treatment (Fig. 4b). Furthermore, those NPR compounds remarkably reduced the number of aggresomes, which contained denatured and misfolded protein aggregates, such as PrPSc, in the cells (Fig. 4c). These results suggest that the candidate NPR compounds that were identified using the original docking system obviously reduced PrPSc in persistently prion-infected cells.
Dose–response of the PrPSc suppressive effects by anti-prion compounds in prion-infected cells. (a) PrPSc inhibitory effects of NPR-130 and NPR-162 compounds were validated using N2a-FK cells treated with each concentration (0, 0.1, 0.5, 1, 5, and 10 μM) for 48 h. PrPSc levels were detected using immunoblotting. (b, c) Localization of PrPSc (b) and aggresomes (c) in N2a-FK cells after 10 μM NPR-130 or NPR-162 treatments for 48 h was analyzed by immunofluorescent staining. PrPSc (green), aggresomes (red), and nuclei (blue) in the cells were detected using a SAF61 antibody, ProteoStat, and DAPI, respectively, and visualized using confocal laser-scanning microscopy. Scale bars represent 10 μm
Effects of Candidate Compounds on PrPC Levels in Prion-Uninfected Cells
To confirm the effect of NPRs on the normal form of PrPC and cell toxicity, we measured PrPC levels and cell viability in prion-uninfected cells after NPR treatment for 48 h. PrPC levels in N2a-58 cells treated with several concentrations of NPR-130 and NPR-162 were similar to the untreated cells (Fig. 5a). Furthermore, the cell viability in the cells treated with those NPRs was similar compared with the cells treated with DMSO only (Fig. 5b). Moreover, the cell viability was not significantly different among the NPRs (Supplementary Fig. 4). These results indicate that NPRs did not have an effect on PrPC levels and cell viability in prion-uninfected cells. Therefore, NPRs may be safe to use as anti-prion drugs.
Effect of PrPC expression in noninfected cells after treatment with the candidate compounds. (a) PrPC levels in N2a-58 cells treated with different concentrations of NPR-130 or NPR-162 compounds (0, 0.1, 0.5, 1, 5, and 10 μM) for 48 h determined using immunoblotting (top). β-Actin was shown as the loading control (middle). Graphs represent the quantitative results of PrPC levels normalized by β-actin (bottom). The data represent 3–5 independent experiments. (b) The living cells after 10 μM treatment of each compound for 48 h were visualized using phase-contrast microscopy (top). Cells were treated with 0.1% DMSO as a vehicle control. The graph indicates cell viability (%). The relative number of living cells was determined after treatment with 10 μM of the indicated compounds for 48 h. Scale bars represent 100 μm. Data are presented as mean ± SEM
Bioassay Using Prion Disease In Vivo Model After Candidate Compound Treatment
To investigate whether candidate compounds exhibited therapeutic effect against prion disease model animals, 4-week-old male mice, inoculated with brain homogenate from terminal phase mice that infected Fukuoka-1 prion, were administrated either compound NPR-130 or NPR-162 three times weekly from immediately post inoculation until the terminal stage of disease. In a bioassay to evaluate whether NPR-130 and NPR-162 compounds have therapeutic effect against prion disease, we discovered that the survival periods of prion-infected mice treated with those compounds were markedly prolonged compared with those of the vehicle treatment group (Fig. 6a). The mice that were intracerebrally inoculated with Fukuoka-1 prion were intraperitoneally administered those compounds at 2 mg/kg/day from 2 days post inoculation until sacrifice. The survival periods of NPR-130-treated mice were significantly longer (291 ± 134 days, n = 10; P = 0.0381, log-rank test) compared with the vehicle mice (189 ± 23 days, n = 9). Moreover, the survival periods of NPR-162-treated mice were also significantly prolonged (258 ± 93 days, n = 11; P = 0.0047, log-rank test) compared with those of the vehicle mice. Intriguingly, some mice treated with the compounds absolutely did not develop prion disease (Fig. 6a). We investigated PrPSc levels in the brain and spleen at 100 d.p.i. as the presymptomatic period and at the terminal phase as the symptomatic period with immunoblotting. NPR-162 treatment significantly suppressed the PrPSc levels in the brain at the terminal phase and in the spleen at both periods (Fig. 6b and Supplementary Fig. 5a). In addition, NPRP-130 treatment suppressed PrPSc levels in the brain and spleen compared with the vehicle treatment, but these results were not significant (Fig. 6b, Supplementary Fig. 5a, b). To evaluate the underlying pathological mechanisms in prion disease, we analyzed the histological changes in the brain at 100 d.p.i., with a particular focus on the thalamus area where critical pathological changes in the brain of prion disease model mice could be observed (Fig. 7). NPR-130 and NPR-162 treatments clearly and significantly suppressed the vacuolar area, the PrPSc-positive area, with a significant reduction of 50% or less than in the vehicle treatment (Fig. 7a, b). Furthermore, to evaluate gliosis levels, we performed immunohistochemical staining with an ionized calcium-binding adaptor molecule-1 (Iba-1) (a marker of microglial activation) and GFAP (a marker of astrocyte activation) antibodies. The treatments with each compound significantly reduced Iba-1 and GFAP-positive cell area in the thalamus (Fig. 7 a, b). In addition to the histological changes in other brain regions, such as the cortex, hippocampus, pons, and cerebellum, NPR-130 and NPR-162 treatments significantly reduced the vacuolar area in the hippocampus and the PrPSc plaques in the pons (Supplementary Fig. 6a–c). Moreover, NPR-162 treatment also significantly suppressed the Iba-1-positive cell area in the cerebellum and the GFAP-positive cell area in the cortex, pons, and cerebellum (Supplementary Fig. 7a–c). NPR-130 treatment also significantly reduced GFAP-positive cell area, but there was no significant difference in Iba-1-positive cell area (Supplementary Fig. 7a–c). These results showed that NPR-130 and NPR-162 inhibited the pathological changes caused by vacuolation, PrPSc plaque formation, and gliosis in the brain after prion infection and delayed prion disease progression. Thus, these compounds might be effective therapeutic drugs for this disease.
Efficacy evaluation of candidate compounds using the prion bioassay system. Analysis of prion pathogenesis in prion-infected mice treated with either NPR-130 or NPR-162 compounds from 2 days post inoculation (d.p.i.). (a) Survival curves indicated candidate compound-treated mice post prion inoculation. Distinct mice in graph are displayed as vehicle (circle: n = 9), NPR-130 (triangle: n = 10), and NPR-162 (square: n = 11). (b) Vehicle and compound-treated mice were sacrificed at 100 d.p.i. and PrPSc levels in the brains were investigated by immunoblotting. PrPSc accumulation was evaluated by quantification of each band intensity. The graphs show the results of vehicle (white), NPR-130 (gray), and NPR-162-treated groups (black). Statistical significances in survival assay of prion-infected mice was determined using the log-rank test, and the one-way analysis of variance followed by the Tukey–Kramer test for multiple comparisons was used for immunoblotting. *P < 0.05 vs vehicle. Data are presented as mean ± SEM
Histological analysis of the brain in prion-infected mice treated with the candidate compounds. (a) Pathological analysis of the vacuolation changes (HE), accumulation of PrPSc, microgliosis (IBA-1), and astrogliosis (GFAP) in the thalamus area of the brain of compound-treated mice at 100 d.p.i. Scale bars represent 50 μm. (b) Those pathological analyses are evaluated by quantifying the vacuolation area, PrPSc area, Iba-1-positive cells, and GFAP-positive cells following immunohistochemistry. In all graphs, white, gray, and black circles indicate vehicle-, NPR-130-, and NPR-162-treated groups, respectively. Statistical significances were determined using the one-way analysis of variance followed by the Tukey–Kramer test for multiple comparison. *P < 0.05 and **P < 0.01 vs vehicle. Data are presented as mean ± SEM
Discussion
Many clinical studies on prion disease have been conducted by researchers around the world. However, there has been no practical application of prion compounds in the development of prion disease drugs until now. Challenges persist in elucidating the mechanism involved in protein structure conversion from normal forms, such as PrPC, to aberrant forms, such as PrPSc, and the real conformation of PrPSc, which is an estimated pathogen of prion disease. In our previous report, we investigated ~ 210,000 compounds using the NUDE/DEGIMA supercomputer system, which was developed as an original docking simulation, and identified candidate compounds expected to have therapeutic effects against prion disease, but the effects were not fully elucidated [8]. Therefore, we again focused on the conformation of PrPC and investigated novel compounds which bind to the PrPC, as well as the stabilization of the protein structure to prevent this conversion [6, 8], and searched for candidate compounds that bind to the “hot spot” from ~ 690,000 compounds. Here, we discovered that 17 of the top 73 compounds exhibited an anti-prion effect that significantly decreased the PrPSc level in prion-infected cells, indicating the hit rate in this study was 23.3% (17 out of 73 compounds), approximately four times higher than that of the previous study results [8]. This indicates that the accuracy of our analysis might have been enhanced by the greater number of compounds in this library and the specificity of the target region of PrPC conformation.
Several compounds exhibited a PrPSc reduction effect in prion-infected cells; however, those compounds never exhibited a strong biding affinity for PrPC in this SPR study (Figs. 1, 3, and 5) because we analyzed the binding affinity between candidate compounds and full-length PrP, but not the restrictive “hot spot” region of PrPC. If we had evaluated the SPR analysis using only the “hot spot” of PrPC, almost all compounds might have shown RU values of higher intensity. Moreover, we would have had to confirm the detailed binding region in PrPC and validate whether the binding specificity of these compounds against PrPC by competition SPR analysis using hot spot binding compounds such as GN8 or NPR analysis to evaluate similarly chemical shift perturbations. Thus, further experiments are needed to clarify the speculation. The potential problem will be validated and investigated in a future manuscript.
In our virtual screening, we calculated the docking simulation using data of human PrP conformation. Thus, the number of compounds that exhibited a binding of more than 10 RU in SPR analysis were mostly cases of human PrP than of mouse PrP (Supplementary Fig. 1). Compounds NPR-130 and NPR-162, in particular, showed a high binding affinity to not only human PrP but also mouse PrP compared with other compounds. The FMO calculations indicated that this high binding affinity was mainly caused by nonpolar interactions (i.e., van der Waals or dispersion interactions). This binding character is very different from that of GN8, in which a polar interaction is a major factor responsible for the molecular interaction with PrPC. Both compounds also showed a therapeutic effect that prolonged the survival periods after prion infection and suppressed typical brain lesions of prion-infected mice before the onset of histopathological analysis (Fig. 7, Supplementary Figs. 6 and 7), indicating that, unlike previous candidate compounds, both compounds could exhibit excellent effects on human prion disease.
Conclusion
In silico drug discovery based on data of protein conformation is a ready and prominent method to enable us to efficiently acquire candidate drugs for many diseases. However, accomplishment of the virtual screening demands reliable and accurate information acquisition of the critical proteins related to the disease. In the studies on prion disease, only conformation data from PrPC of not full-length but restricted epitope has been reported so far; meaning that nobody has a true conformation data of human PrPC. If docking simulation was calculated by the DEGIMA system using precise information of a full-length PrPC, we might be able to obtain better compounds than NPR-130 and NPR-162 in this study. Furthermore, if we will acquire the true conformation data of both PrPC and PrPSc, we might be able to develop a superior and practical compound as a sole therapeutic drug for prion disease.
Recently, we already reported that using our original virtual screening, we developed effective compounds that targeted the PA–PB1 complex of influenza virus [38]. It is indicated that our original in silico screening system using a structure-based drug discovery is excellent for rapidly examining candidate compounds that bind to the distinctive pathogenesis-related target protein in any disease and is likely to have therapeutic effect against the disease. In the future, in silico screening using STDD will be used more extensively, and this has a promising potential.
Our study findings revealed that NPR-130 and NPR-162 compounds had an unprecedented therapeutic effect that prodigiously prolonged survival periods in prion-infected mice. Thus, both compounds are proven to be overwhelmingly close to being practical drugs and might treat prion disease compared with candidate compounds that have been reported until now. Collectively, these results prove the outstanding nature of our drug discovery project strategy.
References
Shyng SL, Lehmann S, Moulder KL, Harris DA. Sulfated glycans stimulate endocytosis of the cellular isoform of the prion protein, PrPC, in cultured cells. J Biol Chem. 1995;270(50):30221-9.
Mange A, Nishida N, Milhavet O, McMahon HE, Casanova D, Lehmann S. Amphotericin B inhibits the generation of the scrapie isoform of the prion protein in infected cultures. Journal of Virology. 2000;74(7):3135-40.
Marella M, Lehmann S, Grassi J, Chabry J. Filipin prevents pathological prion protein accumulation by reducing endocytosis and inducing cellular PrP release. The Journal of Biological Chemistry. 2002;277(28):25457-64.
Kiachopoulos S, Heske J, Tatzelt J, Winklhofer KF. Misfolding of the prion protein at the plasma membrane induces endocytosis, intracellular retention and degradation. Traffic. 2004;5(6):426-36.
Homma T, Ishibashi D, Nakagaki T, Fuse T, Mori T, Satoh K, et al. Ubiquitin-specific protease 14 modulates degradation of cellular prion protein. Scientific Reports. 2015;5:11028.
Kuwata K, Nishida N, Matsumoto T, Kamatari YO, Hosokawa-Muto J, Kodama K, et al. Hot spots in prion protein for pathogenic conversion. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(29):11921-6.
Yamaguchi K, Kamatari YO, Ono F, Shibata H, Fuse T, Elhelaly AE, et al. A designer molecular chaperone against transmissible spongiform encephalopathy slows disease progression in mice and macaques. Nat Biomed Eng. 2019;3(3):206-19.
Ishibashi D, Nakagaki T, Ishikawa T, Atarashi R, Watanabe K, Cruz FA, et al. Structure-based drug discovery for prion disease using a novel binding simulation. EBioMedicine. 2016;9:238-49.
Peretz D, Williamson RA, Kaneko K, Vergara J, Leclerc E, Schmitt-Ulms G, et al. Antibodies inhibit prion propagation and clear cell cultures of prion infectivity. Nature. 2001;412(6848):739-43.
Enari M, Flechsig E, Weissmann C. Scrapie prion protein accumulation by scrapie-infected neuroblastoma cells abrogated by exposure to a prion protein antibody. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(16):9295-9.
White AR, Enever P, Tayebi M, Mushens R, Linehan J, Brandner S, et al. Monoclonal antibodies inhibit prion replication and delay the development of prion disease. Nature. 2003;422(6927):80-3.
Soto C, Kascsak RJ, Saborio GP, Aucouturier P, Wisniewski T, Prelli F, et al. Reversion of prion protein conformational changes by synthetic beta-sheet breaker peptides. Lancet. 2000;355(9199):192-7.
Supattapone S, Wille H, Uyechi L, Safar J, Tremblay P, Szoka FC, et al. Branched polyamines cure prion-infected neuroblastoma cells. J Virol. 2001;75(7):3453-61.
Forloni G, Salmona M, Marcon G, Tagliavini F. Tetracyclines and prion infectivity. Infect Disord Drug Targets. 2009;9(1):23-30.
Forloni G, Iussich S, Awan T, Colombo L, Angeretti N, Girola L, et al. Tetracyclines affect prion infectivity. Proc Natl Acad Sci U S A. 2002;99(16):10849-54.
Chabry J, Priola SA, Wehrly K, Nishio J, Hope J, Chesebro B. Species-independent inhibition of abnormal prion protein (PrP) formation by a peptide containing a conserved PrP sequence. J Virol. 1999;73(8):6245-50.
Ma J, Lindquist S. Conversion of PrP to a self-perpetuating PrPSc-like conformation in the cytosol. Science. 2002;298(5599):1785-8.
Heiseke A, Aguib Y, Riemer C, Baier M, Schatzl HM. Lithium induces clearance of protease resistant prion protein in prion-infected cells by induction of autophagy. Journal of Neurochemistry. 2009;109(1):25-34.
Aguib Y, Heiseke A, Gilch S, Riemer C, Baier M, Schatzl HM, et al. Autophagy induction by trehalose counteracts cellular prion infection. Autophagy. 2009;5(3):361-9.
Ishibashi D, Homma T, Nakagaki T, Fuse T, Sano K, Takatsuki H, et al. Strain-dependent effect of macroautophagy on abnormally folded prion protein degradation in infected neuronal cells. PloS One. 2015;10(9):e0137958.
Nakagaki T, Satoh K, Ishibashi D, Fuse T, Sano K, Kamatari YO, et al. FK506 reduces abnormal prion protein through the activation of autolysosomal degradation and prolongs survival in prion-infected mice. Autophagy. 2013;9(9):1386-94.
Marzo L, Marijanovic Z, Browman D, Chamoun Z, Caputo A, Zurzolo C. 4-Hydroxytamoxifen leads to PrPSc clearance by conveying both PrPC and PrPSc to lysosomes independently of autophagy. Journal of Cell Science. 2013;126(Pt 6):1345-54.
Doh-ura K, Ishikawa K, Murakami-Kubo I, Sasaki K, Mohri S, Race R, et al. Treatment of transmissible spongiform encephalopathy by intraventricular drug infusion in animal models. Journal of Virology. 2004;78(10):4999-5006.
Demaimay R, Adjou KT, Beringue V, Demart S, Lasmezas CI, Deslys JP, et al. Late treatment with polyene antibiotics can prolong the survival time of scrapie-infected animals. J Virol. 1997;71(12):9685-9.
Masullo C, Macchi G, Xi YG, Pocchiari M. Failure to ameliorate Creutzfeldt-Jakob disease with amphotericin B therapy. J Infect Dis. 1992;165(4):784-5.
Tsuboi Y, Doh-Ura K, Yamada T. Continuous intraventricular infusion of pentosan polysulfate: clinical trial against prion diseases. Neuropathology : official journal of the Japanese Society of Neuropathology. 2009;29(5):632-6.
Haik S, Marcon G, Mallet A, Tettamanti M, Welaratne A, Giaccone G, et al. Doxycycline in Creutzfeldt-Jakob disease: a phase 2, randomised, double-blind, placebo-controlled trial. The Lancet Neurology. 2014;13(2):150-8.
Anderson AC. The process of structure-based drug design. Chem Biol. 2003;10(9):787-97.
Wlodawer A, Vondrasek J. Inhibitors of HIV-1 protease: a major success of structure-assisted drug design. Annu Rev Biophys Biomol Struct. 1998;27:249-84.
Clark DE. What has computer-aided molecular design ever done for drug discovery? Expert Opin Drug Discov. 2006;1(2):103-10.
Marrakchi H, Laneelle G, Quemard A. InhA, a target of the antituberculous drug isoniazid, is involved in a mycobacterial fatty acid elongation system, FAS-II. Microbiology. 2000;146 (Pt 2):289-96.
Dadashpour S, Tuylu Kucukkilinc T, Unsal Tan O, Ozadali K, Irannejad H, Emami S. Design, synthesis and in vitro study of 5,6-diaryl-1,2,4-triazine-3-ylthioacetate derivatives as COX-2 and beta-amyloid aggregation inhibitors. Arch Pharm (Weinheim). 2015;348(3):179-87.
Miller Z, Kim KS, Lee DM, Kasam V, Baek SE, Lee KH, et al. Proteasome inhibitors with pyrazole scaffolds from structure-based virtual screening. J Med Chem. 2015;58(4):2036-41.
Matsuno K, Masuda Y, Uehara Y, Sato H, Muroya A, Takahashi O, et al. Identification of a new series of STAT3 inhibitors by virtual screening. ACS Med Chem Lett. 2010;1(8):371-5.
Grover S, Apushkin MA, Fishman GA. Topical dorzolamide for the treatment of cystoid macular edema in patients with retinitis pigmentosa. Am J Ophthalmol. 2006;141(5):850-8.
Perrier V, Wallace AC, Kaneko K, Safar J, Prusiner SB, Cohen FE. Mimicking dominant negative inhibition of prion replication through structure-based drug design. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(11):6073-8.
Hyeon JW, Choi J, Kim SY, Govindaraj RG, Jam Hwang K, Lee YS, et al. Discovery of novel anti-prion compounds using in silico and in vitro approaches. Scientific Reports. 2015;5:14944.
Watanabe K, Ishikawa T, Otaki H, Mizuta S, Hamada T, Nakagaki T, et al. Structure-based drug discovery for combating influenza virus by targeting the PA-PB1 interaction. Sci Rep. 2017;7(1):9500.
Biljan I, Giachin G, Ilc G, Zhukov I, Plavec J, Legname G. Structural basis for the protective effect of the human prion protein carrying the dominant-negative E219K polymorphism. The Biochemical Journal. 2012;446(2):243-51.
Yamamoto N, Kuwata K. Regulating the conformation of prion protein through ligand binding. J Phys Chem B. 2009;113(39):12853-6.
Kitaura K, Sawai T, Asada Y, Nakano T, Uebayasi M. Pair interaction molecular orbital method: an approximate computational method for molecular interactions. Chemical Physics Letters. 1999;312(2-4):319-24.
Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11(8):3696-713.
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. Journal of Computational Chemistry. 2004;25(9):1157-74.
Ishikawa T, Kuwata K. Fragment molecular orbital calculation using the RI-MP2 method. Chemical Physics Letters. 2009;474(1-3):195-98.
Dunning TH. Gaussian basis sets for use in correlated molecular calculations. I. The atoms boron through neon and hydrogen. The Journal of Chemical Physics. 1989;90(2):1007-23.
Ishikawa T, Ishikura T, Kuwata K. Theoretical study of the prion protein based on the fragment molecular orbital method. Journal of Computational Chemistry. 2009;30(16):2594-601.
Atarashi R, Satoh K, Sano K, Fuse T, Yamaguchi N, Ishibashi D, et al. Ultrasensitive human prion detection in cerebrospinal fluid by real-time quaking-induced conversion. Nature Medicine. 2011;17(2):175-8.
Ishibashi D, Yamanaka H, Mori T, Yamaguchi N, Yamaguchi Y, Nishida N, et al. Antigenic mimicry-mediated anti-prion effects induced by bacterial enzyme succinylarginine dihydrolase in mice. Vaccine. 2011;29(50):9321-8.
Homma T, Ishibashi D, Nakagaki T, Satoh K, Sano K, Atarashi R, et al. Increased expression of p62/SQSTM1 in prion diseases and its association with pathogenic prion protein. Scientific Reports. 2014;4:4504.
Ishibashi D, Atarashi R, Fuse T, Nakagaki T, Yamaguchi N, Satoh K, et al. Protective role of interferon regulatory factor 3-mediated signaling against prion infection. Journal of Virology. 2012;86(9):4947-55.
Ishibashi D, Homma T, Nakagaki T, Fuse T, Sano K, Satoh K, et al. Type I interferon protects neurons from prions in in vivo models. Brain. 2019;142(4):1035-50.
Acknowledgments:
We thank Yuzuru and Yukiko Miyazaki-Hirota from Nagasaki University, for the helpful discussions and critical assessment of the manuscript and Atsuko Matsuo, Hanako Nakayama, Marie Yamaguchi, and Megumi Tanaka for the technical assistance. We thank Enago (www.enago.jp) for the English language review.
This work was supported in part by the a grant from a Grant-in-Aid of the Research Committee of Prion Disease and Slow Virus Infection from the Ministry of Health, Labour and Welfare of Japan (N.N.); a Grant-in-Aid of the Research Committee of Molecular Pathogenesis and Therapies for Prion Disease and Slow Virus Infection, the Practical Research Project for Rare and Intractable Disease from the Japan Agency for Medical Research and Development, AMED (D.I.); a grant from the Takeda Science Foundation (N.N. and D.I.); a grant from the Japan Intractable Disease Research Foundation (D.I.); a Grant-in-Aid from the Tokyo Biochemical Research Foundation (D.I.); a grant provided by the YOKOYAMA Foundation for Clinical Pharmacology (Grant No. YRY1502) (D.I.); a grant provided by the Ichiro Kanehara Foundation (D.I.); a grant provided by the Mochida Memorial Foundation for Medical and Pharmaceutical Research (D.I.); a grant provided by the Waksman Foundation of Japan Inc. (D.I.); and a grant provided by the Center for Clinical and Translational Research of Kyushu University Hospital (D.I.).
Author Contribution Statement
D.I., T.I., S.M., H.T., T.N., and T.H. coordinated and performed the entire project. T.I. and N.N. supervised and discussed the data. D.I., T.I., and N.N. wrote the manuscript. D.I. and N.N. revised the manuscript.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Availability of Data and Materials
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary material.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Consent for Publication
This manuscript does not contain data from any individual person.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ishibashi, D., Ishikawa, T., Mizuta, S. et al. Novel Compounds Identified by Structure-Based Prion Disease Drug Discovery Using In Silico Screening Delay the Progression of an Illness in Prion-Infected Mice. Neurotherapeutics 17, 1836–1849 (2020). https://doi.org/10.1007/s13311-020-00903-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13311-020-00903-9