Abstract
The study presents an in silico identification of poly (cis-1,4-isoprene) cleaving enzymes, viz., RoxA and RoxB in bacteria, followed by their functional and evolutionary exploration using comparative genomics. The orthologs of these proteins were found to be restricted to Gram-negative beta-, gamma-, and delta-proteobacteria. Toward the evolutionary propagation, the RoxA and RoxB genes were predicted to have evolved via a common interclass route of horizontal gene transfer in the phylum Proteobacteria (delta → gamma → beta). Besides, recombination, mutation, and gene conversion were also detected in both the genes leading to their diversification. Further, the differential selective pressure is predicted to be operating on entire RoxA and RoxB genes such that the former is diversifying further, whereas the latter is evolving to reduce its genetic diversity. However, the structurally and functionally important sites/residues of these genes were found to be preventing changes implying their evolutionary conservation. Further, the phylogenetic analysis demonstrated a sharp split between the RoxA and RoxB orthologs and indicated the emergence of their variant as another type of putative rubber oxygenase (RoxC) in the class Gammaproteobacteria. A detailed in silico analysis of the signature motifs and residues of Rox sequences exhibited important differences as well as similarities among the RoxA, RoxB, and putative RoxC sequences. Although RoxC appears to be a hybrid of RoxA and RoxB, the signature motifs and residues of RoxC are more similar to RoxB.





Similar content being viewed by others
References
Arber W (2014) Horizontal gene transfer among bacteria and its role in biological evolution. Life (Basel) 4:217–224
Bendtsen JD, Kiemer L, Fausbøll A, Brunak S (2005) Non-classical protein secretion in bacteria. BMC Microbiol 5:58
Betrán E, Rozas J, Navarro A, Barbadilla A (1997) The estimation of the number and the length distribution of gene conversion tracts from population DNA sequence data. Genetics 146:89–99
Birke J, Jendrossek D (2014) Rubber oxygenase (RoxA) and latex clearing protein (Lcp) cleave rubber to different products and use different cleavage mechanisms. Appl Environ Microbiol 80:5012–5020
Birke J, Hambsch N, Schmitt G, Altenbuchner J, Jendrossek D (2012) Phe317 is essential for rubber oxygenase RoxA activity. Appl Environ Microbiol 78:7876–7883
Birke J, Röther W, Schmitt G, Jendrossek D (2013) Functional identification of rubber oxygenase (RoxA) in soil and marine myxobacteria. Appl Environ Microbiol 79:6391–6399
Birke J, Röther W, Jendrossek D (2017) RoxB is a novel type of rubber oxygenase that combines properties of rubber oxygenase RoxA and latex clearing protein (Lcp). Appl Environ Microbiol 83:e00721–e1717
Birke J, Röther W, Jendrossek D (2018) Rhizobacter gummiphilus NS21 has two rubber oxygenases (RoxA and RoxB) acting synergistically in rubber utilisation. Appl Microbiol Biotechnol 102:10245–10257
Boc A, Makarenkov VJ (2011) Towards an accurate identification of mosaic genes and partial horizontal gene transfers. Nucleic Acids Res 39:e144–e144
Boc A, Philippe H, Makarenkov V (2010) Inferring and validating horizontal gene transfer events using bipartition dissimilarity. Syst Biol 59:195–211
Boc A, Diallo AB, Makarenkov V (2012) T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res 40:W573–W579
Boni MF, Posada D, Feldman MW (2007) An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 176:1035–1047
Braaz R, Armbruster W, Jendrossek D (2005) Heme-dependent rubber oxygenase RoxA of Xanthomonas sp. cleaves the carbon backbone of poly (cis-1, 4-isoprene) by a dioxygenase mechanism. Apwbiol 71:2473–2478
Bruen TC, Philippe H, Bryant D (2006) A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665–2681
Bryant J, Chewapreecha C, Bentley SD (2012) Developing insights into the mechanisms of evolution of bacterial pathogens from whole-genome sequences. Future Microbiol 7:1283–1296
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K et al (2009) BLAST+: architecture and applications. BMC Bioinformatics 10:421
Costa J, Teixeira PG, d'Avó AF, Júnior CS, Veríssimo A (2014) Intragenic recombination has a critical role on the evolution of Legionella pneumophila virulence-related effector sidJ. PLoS One 9:e109840
Darmon E, Leach DR (2014) Bacterial genome instability. Microbiol Mol Biol Rev 78:1–39
Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM et al (2011) T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res 39:W13–W17
Durbin R, Eddy SR, Krogh A, Mitchison G (1998) Biological sequence analysis: probabilistic models of proteins and nucleic acids. Cambridge University Press, Cambridge
Eddy SR (2011) Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Francino MP (2012) The ecology of bacterial genes and the survival of the new. Int J Evol Biol 2012:394026
Gibbs MJ, Armstrong JS, Gibbs AJ (2000) Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16:573–582
Grove A (2010) Functional evolution of bacterial histone-like HU proteins. Curr Issues Mol Biol 13:1–12
Gruen DS, Wolfe JM, Fournier GP (2019) Paleozoic diversification of terrestrial chitin-degrading bacterial lineages. BMC Evol Biol 19:34
Gupta RS (1998) Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol Mol Biol Rev 62:1435–1491
Hagblom P, Segal E, Billyard E, So M (1985) Intragenic recombination leads to pilus antigenic variation in Neisseria gonorrhoeae. Nature 315:156–158
Ho-Huu J, Ronfort J, De Mita S, Bataillon T, Hochu I, Weber A et al (2012) Contrasted patterns of selective pressure in three recent paralogous gene pairs in the Medicago genus (L.). BMC Evol Biol 12:195
Hu T, Banzhaf W (2008) Nonsynonymous to Synonymous Substitution Ratio Ka/Ks: measurement for rate of evolution in evolutionary computation. In: Proceedings of 10th International Conference on Parallel Problem Solving from Nature (PPSN X). Springer-Verlag GmbH, Berlin, Germany, pp 448–457.
Hurst LD (2002) The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet 18:486–487
Huson DH, Bryant D (2005) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119
Jendrossek D, Birke J (2019) Rubber oxygenases. Appl Microbiol Biotechnol 103:125–142
Jendrossek D, Reinhardt S (2003) Sequence analysis of a gene product synthesized by Xanthomonas sp. during growth on natural rubber latex. FEMS Microbiol Lett 224:61–65
Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C et al (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240
Juárez-Vázquez AL, Edirisinghe JN, Verduzco-Castro EA, Michalska K, Wu C, Noda-García L, Babnigg G, Endres M, Medina-Ruíz S, Santoyo-Flores J, Carrillo-Tripp M, Ton-That H, Joachimiak A, Henry CS, Barona-Gómez F (2017) Evolution of substrate specificity in a retained enzyme driven by gene loss. Elife 6:e22679
Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A (2003) Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci 12:1652–1662
Kahsay RY, Gao G, Liao L (2005) An improved hidden Markov model for transmembrane protein detection and topology prediction and its applications to complete genomes. Bioinformatics 21:1853–1858
Kasai D, Imai S, Asano S, Tabata M, Iijima S, Kamimura N et al (2017) Identification of natural rubber degradation gene in Rhizobacter gummiphilus NS21. Biosci Biotechnol Biochem 81:614–620
Katoh K, Standley DM (2013) MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Librado P, Rozas J (2009) DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Mansai SP, Kado T, Innan H (2011) The rate and tract length of gene conversion between duplicated genes. Genes 2:313–331
Martin D, Rybicki E (2000) RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562–563
Martin D, Posada D, Crandall K, Williamson C (2005) A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retrov 21:98–102
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003
McVean G, Awadalla P, Fearnhead P (2002) A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics 160:1231–1241
Na S-I, Kim YO, Yoon S-H, Ha S-m, Baek I, Chun J (2018) UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol 56:281–285
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426
Nielsen KM, Bøhn T, Townsend JP (2014) Detecting rare gene transfer events in bacterial populations. Front Microbiol 4:415
Noor S, Changey F, Oakeshott JG, Scott C, Martin-Laurent F (2014) Ongoing functional evolution of the bacterial atrazine chlorohydrolase AtzA. Biodegradation 25:21–30
Okonechnikov K, Golosova O, Fursov M, Team U (2012) Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167
Padidam M, Sawyer S, Fauquet CM (1999) Possible emergence of new geminiviruses by frequent recombination. Virology 265:218–225
Paulsson J, El Karoui M, Lindell M, Hughes D (2017) The processive kinetics of gene conversion in bacteria. Mol Microbiol 104:752–760
Perry AJ, Ho BK (2013) Inmembrane, a bioinformatic workflow for annotation of bacterial cell-surface proteomes. Source Code Biol Med 8:9
Perry JA, Wright GD (2014) Forces shaping the antibiotic resistome. BioEssays 36:1179–1184
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786
Pond SLK, Frost SD (2005) Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533
Pond SLK, Frost SDW, Muse SV (2005) HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679
Pond SLK, Posada D, Gravenor MB, Woelk CH, Frost SDW (2006) GARD: a genetic algorithm for recombination detection. Bioinformatics 22:3096–3098
Posada D (2002) Evaluation of methods for detecting recombination from DNA sequences: empirical data. Mol Biol Evol 19:708–717
Price MN, Dehal PS, Arkin AP (2010) FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490
Rambaut A (2014) FigTree v1.4.2, A graphical viewer of phylogenetic trees. https://tree.bio.ed.ac.uk/software/figtree Accessed 8 March 2018.
Robinson DR, Foulds LR (1981) Comparison of phylogenetic trees. Math Biosci 53:131–147
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schmitt G, Birke J, Jendrossek D (2019) Towards the understanding of the enzymatic cleavage of polyisoprene by the dihaem-dioxygenase RoxA. AMB Express 9:166
Seidel J, Schmitt G, Hoffmann M, Jendrossek D, Einsle O (2013) Structure of the processive rubber oxygenase RoxA from Xanthomonas sp. Proc Natl Acad Sci USA 110:13833–13838
Shapiro BJ, Alm EJ (2008) Comparing patterns of natural selection across species using selective signatures. PLoS Genet 4:e23
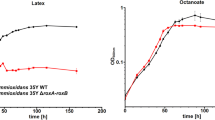
Sharma V, Siedenburg G, Birke J, Mobeen F, Jendrossek D, Prakash T (2018) Metabolic and taxonomic insights into the Gram-negative natural rubber degrading bacterium Steroidobacter cummioxidans sp. nov., strain 35Y. PLoS One 13:0197448
Smith JM (1992) Analyzing the mosaic structure of genes. J Mol Evol 34:126–129
Suyama M, Torrents D, Bork P (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34:W609–W612
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA 101:11030–11035
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tsuchii A, Takeda K (1990) Rubber-degrading enzyme from a bacterial culture. Appl Environ Microbiol 56:269–274
Van Elsas JD, Turner S, Bailey MJ (2003) Horizontal gene transfer in the phytosphere. New Phytol 157:525–537
Van Zee JP, Schlueter JA, Schlueter S, Dixon P, Sierra CA, Hill CA (2016) Paralog analyses reveal gene duplication events and genes under positive selection in Ixodes scapularis and other ixodid ticks. BMC Genomics 17:241
Wachter J, Hill S (2016) Positive selection pressure drives variation on the surface-exposed variable proteins of the pathogenic Neisseria. PLoS One 11:e0161348
Zhang Z, Li J, Zhao X-Q, Wang J, Wong GK-S, Yu J (2006) KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genom Proteom Bioinf 4:259–263
Zhang YZ, Li Y, Xie BB, Chen XL, Yao QQ, Zhang XY et al (2016) Nascent genomic evolution and allopatric speciation of Myroides profundi D25 in its transition from Land to Ocean. mBio 7:e01946–e11915
Zuckerkandl E, Pauling L (1965) Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ (eds) Evolving genes and proteins. Academic Press, New York, pp 97–166
Acknowledgements
TP acknowledges IIT Mandi for financial and technical support. VS acknowledges the Ministry of Human Resource Development (MHRD), India, for providing the research fellowship.
Funding
This research received no external funding.
Author information
Authors and Affiliations
Contributions
TP and VS conceived or designed the study, TP and VS performed research, TP, VS, and FM analyzed data, TP and VS wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sharma, V., Mobeen, F. & Prakash, T. In silico functional and evolutionary analyses of rubber oxygenases (RoxA and RoxB). 3 Biotech 10, 376 (2020). https://doi.org/10.1007/s13205-020-02371-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02371-6