Antibacterial and Cytotoxic Bridged and Ring Cleavage Angucyclinones From a Marine Streptomyces sp
- 1School of Pharmacy, Binzhou Medical University, Yantai, China
- 2College of Life Sciences, Yantai University, Yantai, China
Chemical investigation of a marine-derived Streptomyces sp. KCB-132, cultivated in liquid ISP2 medium, had led to the discovery of three C-ring cleavage angucyclinone N-heterocycles, pratensilins A–C, with a novel spiro indolinone-naphthofuran skeleton. Addition of 50 μM LaCl3 to the same medium and subsequent chemical analysis of this strain returned a new member of this rare class, pratensilin D (1), along with two new angucyclinone derivatives, featuring ether-bridged (2) and A-ring cleavage (3) structural properties. Their structures and absolute configurations were assigned by spectroscopic analysis, single-crystal X-ray diffractions, and equivalent circulating density (ECD) calculations. (+)- and (–)-1, a pair of enantiomeric nitrogen-containing angucyclinones, exhibited different strengths of antibacterial and cytotoxic activities.
Introduction
Angucyclines and angucyclinones (sugarless) represent the largest family of type II polyketide synthase (PKS)-engineered natural products (Rohr and Thiericke, 1992; Krohn and Rohr, 1997; Kharel et al., 2012), which share a characteristic tetracyclic benz[a]anthracene core and exhibit a wide range of biological activities (Shaaban et al., 2012; Ma et al., 2015; Xie et al., 2016; Yixizhuoma et al., 2017; Liu et al., 2019; Wu et al., 2019). Actinobacteria are the exclusive producers of angucyclines and angucyclinones that have been proven to be a prolific source of antibiotics for the production of 45% of all reported microbial active metabolites (Bérdy, 2005; Arens et al., 2013). However, only a few such metabolites are detected in the laboratory due to conditional or low production of most metabolites biosynthesized by cryptic gene clusters (Helge et al., 2002; Demain and Sanchez, 2009; Newman and Cragg, 2012; Bethany and Mohammad, 2017). Rare earth, such as scandium, was reported to be an effective factor that induces or stimulates the production of secondary metabolites (Kawai et al., 2007).
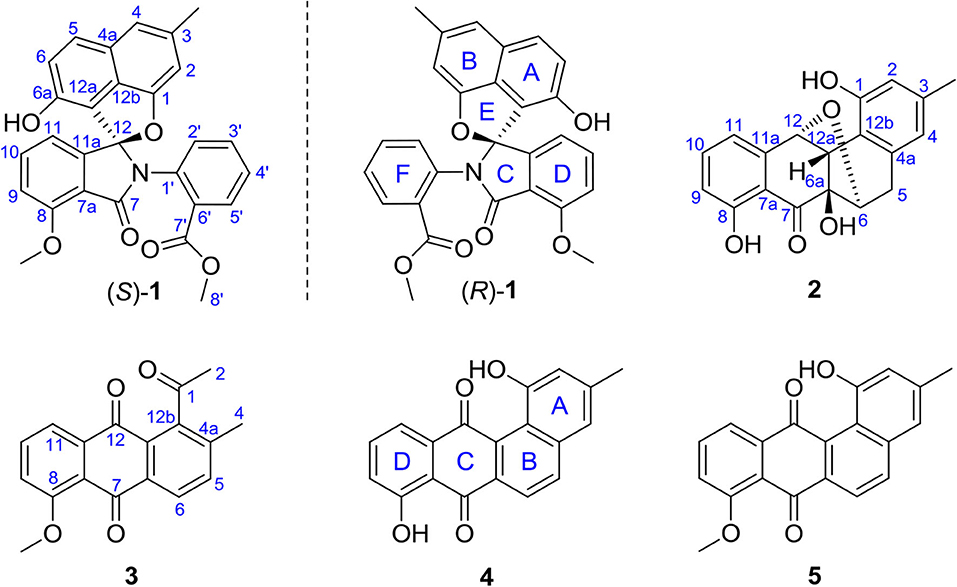
Our previous chemical analysis of the metabolites from marine actinomycete Streptomyces pratensis KCB-132 have led to the discovery of three pairs of unprecedented ring C-modified angucyclinone N-heterocycles, (+)- and (–)-pratensilins A–C. Addition of the rare earth, lanthanum, to the liquid ISP2 medium and repeated purification of the organic extract of this strain resulted in the isolation of two new ring cleavage angucyclinones, named (±)-pratensilin D (1) and pratensinon A (3), and a new ether-bridged analog, kiamycin E (2), together with two known angucyclinones tetrangulol (4) and 8-O-methyltetrangulol (5) (Figure 1). Herein, we report the isolation, structure characterization, and biological activity of compounds 1–3, and a plausible biogenetic pathway for 1 and 2 is also proposed.
Results and Discussion
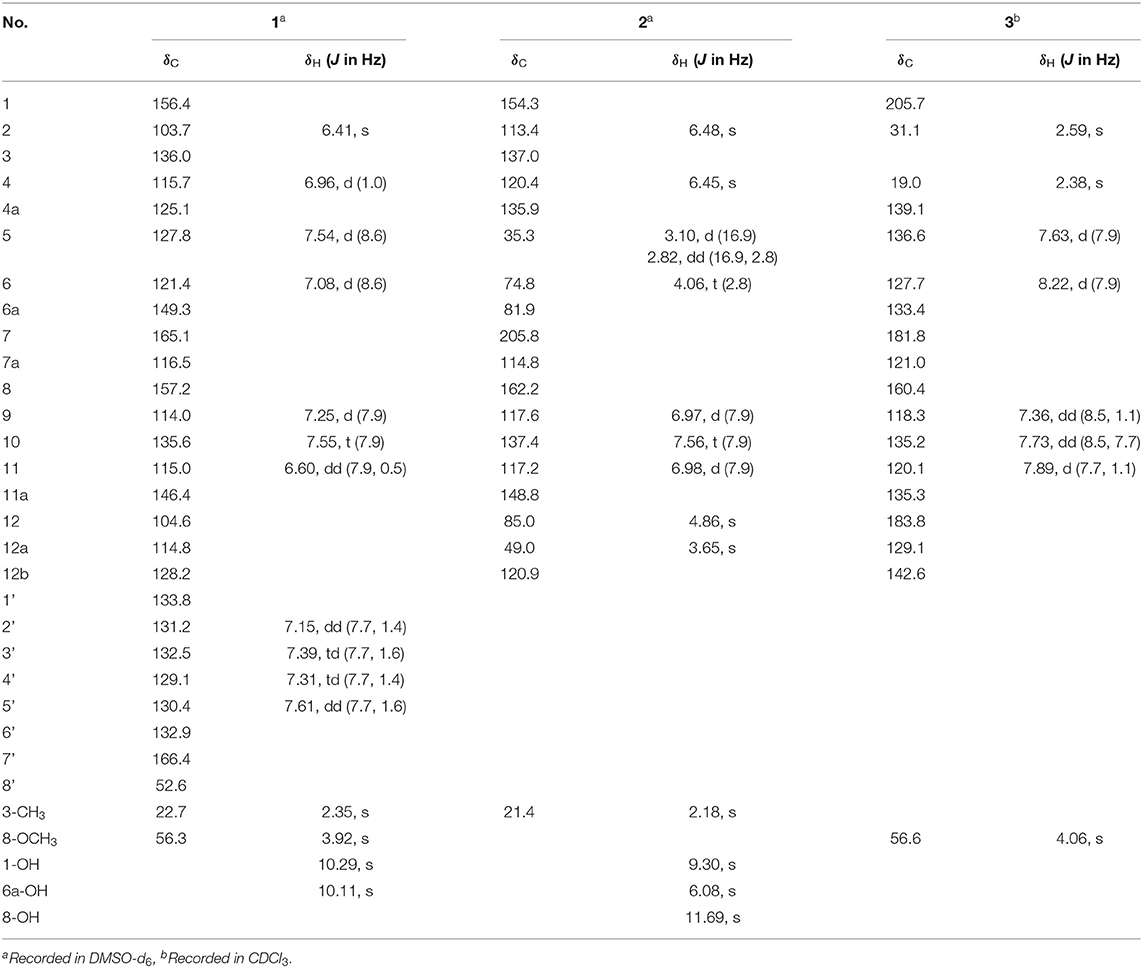
Compound 1 was obtained as a colorless solid. Its molecular formula of C28H21O6N was established from the positive-ion HRESIMS peak at m/z 468.14418 [M + H]+ (calcd for C28H22O6N, 468.14471). The 1H, 13C, and heteronuclear single quantum coherence (HSQC) NMR spectra of 1 displayed one tertiary methyl at δ 2.35, two methoxy groups at δ 3.75 and 3.97, and 11 aromatic methines (δ 7.61, 7.55, 7.54, 7.39, 7.31, 7.25, 7.15, 7.08, 6.96, 6.60, and 6.41), as well as one exchangeable proton at δ 10.11. The 13C NMR spectrum indicated the presence of two carbonyls at δ 165.1 and 166.4 and 12 quaternary carbons between δ 103.7 and 157.2 (Table 1). Interpretation of the 1H-1H COrrelation SpectroscopY (COSY) spectrum permitted three isolated fragments to be established. The first fragment, a 1,2,3-trisubstituted benzene ring (ring D) was assigned by 1H-1H COSY cross-peaks for H-9/H-10/H-11 along with heteronuclear multiple bond correlations (HMBCs) from H-9 to C-7a and from H-10 to C-8 and C-11a. Next, 1H-1H COSY cross-peaks for H-2′/H-3′/H-4′ and H-5′ along with HMBC couplings from H-2′ to C-6′ and from H-5′ to C-1′ allowed the constitution of a second benzene ring (ring F). The last fragment was established based on 1H-1H COSY cross-signals between H-5 and H-6 in combination with HMBCs from H-5 to C-6a and C-12b and from H-6 to C-4a and C-12a, resulting in a third benzene ring as ring B, which shared double bond Δ4a,12b with a methyl benzene moiety (ring A) to fuse into a naphthalene system, as indicated by HMBCs from H-2 to C-1 and C-12b, from 3-CH3 to C-2, C-3, and C-4, and from H-4 to C-4a (Figure 2A).
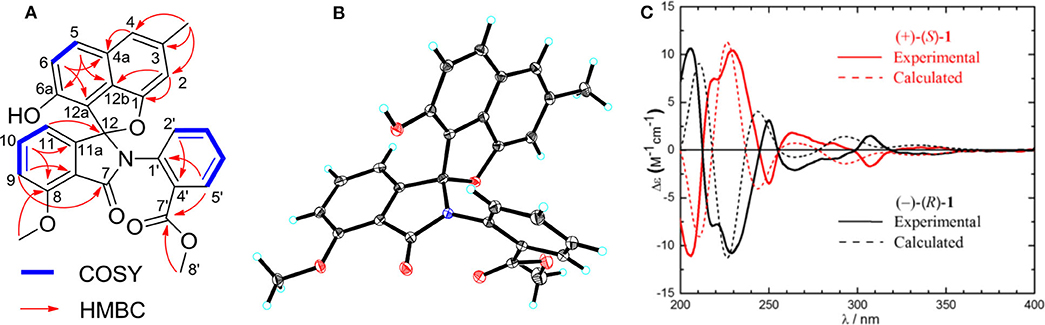
Figure 2. (A) Key 1H-1H COrrelation SpectroscopY (COSY) and heteronuclear multiple bond correlations (HMBCs) of 1. (B) X-ray crystal structure of (–)-1. (C) Experimental and calculated equivalent circulating density (ECD) spectra for (+)-(S)-1 and (–)-(R)-1.
In addition, key HMBCs from H-9 to the carbonyl carbon C-7 (δ 165.1) and from H-11 to the deshielded carbon C-12 (δ 104.6) extended ring D to C-7 and C-12; these two carbons had to be connected by an oxygen or a nitrogen atom to form either an isobenzofuran-1-one or an isoindolin-1-one ring system in line with both the chemical shifts and the molecular composition. However, both ring systems had appeared in the structures of C-ring cleavage angucyclinones produced by the same strain, previously. Moreover, no obvious correlations were observed between ring F and other ring systems in the 1H-1H COSY and HMBC spectra, which made it problematic to assign the complete structure by spectroscopic analysis.
Finally, the structure of 1 was elucidated by X-ray crystallographic analysis (Figure 2B). Slow evaporation of a MeOH/CH2Cl2 solution of 1 provided suitable crystals. X-ray experiment revealed that 1 was composed of a methyl benzoate moiety attached to a spiro indolinone-naphthofuran core skeleton. Additionally, the X-ray data exhibited a centrosymmetric space group C12/c1, supporting a racemic nature. Subsequent optical resolution of 1 was achieved by high-performance liquid chromatography (HPLC) equipped with a chiral column to give (+)-1 and (–)-1. Based on the analogy to pratensilins A–C, we suggest that the absolute configurations for the two isomers can be assigned as (+)-S-1 and (–)-R-1 for a biogenetic reasoning. As in pratensilins A–C, the (S)-enantiomers have positive optical rotations, while the (R)-enantiomers share consistently negative optical rotations (Zhang et al., 2017). This assignment was also confirmed by equivalent circulating density (ECD) calculations using the same ECD computational approach as for pratensilins A–C. In fact, the CD spectra of (+)-1 and (–)-1 reproduced well with those of pratensilins A–C apart from a systematic wavelength shift (Figure 2C). Herein, we suggest to name pratensilin D for compound 1.
Compound 2 was isolated as a pale-yellow solid. High-resolution electrospray ionization mass spectrometry (HRESIMS) provided a molecular formula of C19H16O6 [m/z 325.10740 [M + H]+, calcd for C19H17O5, 325.10760] requiring 12 degrees of unsaturation. The 13C and distortionless enhancement by polarization transfer (DEPT) NMR spectra exhibited 19 carbon signals for one methyl, one methylene, eight methines including five aromatic methines, one carbonyl carbon, and eight quaternary carbons, of which seven are sp2 aromatic carbons (Table 1). Since the aromatic carbons and the carbonyl accounted for 7 of the 12 degrees of unsaturation, compound 2 must be pentacyclic. In the 1H NMR spectrum, the characteristic signals for o-coupled protons at δ 6.97 (H-9), δ 7.56 (H-10), and δ 6.98 (H-11) indicated the presence of a 1, 2, 3-trisubstituted benzene ring D, while a typical olefinic methyl resonance at δ 2.18 (3-CH3), which showed long-range couplings with two aromatic singlets at δ 6.48 (H-2) and δ 6.45 (H-4), was assigned to benzene ring A as observed in 1 (Figure 1). An additional 1H-1H COSY cross-peak between H2-5 and H-6 along with HMBCs from H2-5 to C-4a, from H-6 to C-6a, and from H-12a to C-6a and C-12b suggested that the cyclohexene ring B is joined to ring A by sharing the double bond between C-4a and C-12b. Furthermore, ring systems A/B and D were linked by ring C (C-7 and C-12) to form an angucyclinone skeleton evidenced by HMBCs from both H-6 and H-9 to the carbonyl carbon C-7 (δ 205.8) and from H-12 to C-11 and C-12a (Figure 3A).
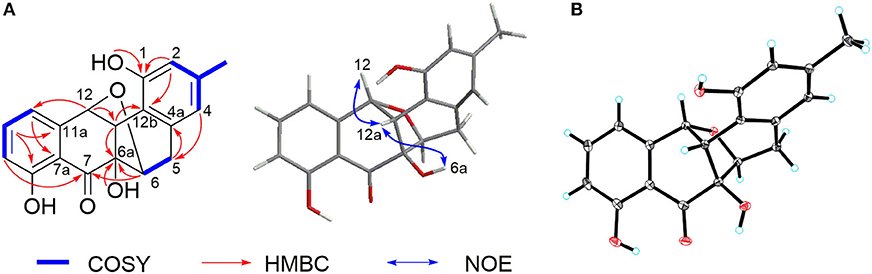
Figure 3. (A) Key 1H-1H COrrelation SpectroscopY (COSY), heteronuclear multiple bond correlation (HMBC), and nuclear Overhauser effect (NOE) correlations of 2. (B) X-ray crystal structure of 2.
Positioning of two hydroxyl groups at C-1 (δ 154.3) and C-6a (δ 81.9) was facilitated by HMBCs from 1-OH to C-1 and from 6a-OH to C-6a, respectively. The remaining phenolic OH group at δ 11.69 had to be attached to the oxygenated aromatic carbon C-8 to fit in the deshielded characteristic of C-8 (δ 162.2). At this point of the structure elucidation, only one oxygen atom was left as defined by the molecular formula, the two downfield methine carbons C-6 and C-12 must be joined by an oxygen bridge in agreement with both the chemical shifts of C-6 (δ 74.8) and C-12 (δ 85.0), completing the pentacyclic requirement of 2, which was further supported by three-bond HMBCs coupling from H-6 to C-12 (and vice versa). Taken together, a rare 6,12-epoxybenz[a] anthracene ring topology was expected; we thus suggest the successive name kiamycin E for 2.
The relative configuration of 2 was tentatively assigned by nuclear Overhauser effect (NOE) correlations (Figure 3A). The NOE contacts of H-12 with H-12a and of H-12a with 6a-OH indicated that H-12, H-12a, and 6-OH have common β-orientations, which require an α-orientation of the bridged C-6/12 bond; consequently, H-6 was defined as β, although no NOE was observed between H-6 and 6a-OH. This assignment was finally confirmed by single-crystal X-ray diffraction experiment (Figure 3B). Additionally, the absolute configuration was assigned as 6S, 6aR, 12R, and 12aS, respectively.
Compound 3 was isolated as a yellow powder. Its molecular formula was assigned to be C18H14O4 based on HRESIMS [m/z 295.09677 [M + H]+, calcd for C18H15O4, 295.09703]. In the 13C NMR spectrum, the typical carbonyl carbon resonances at δ 181.8 and 183.8 indicated the presence of an anthraquinone skeleton. A comparison of the 1H and 13C NMR spectra of 3 and the known derivative 4 revealed superimposable resonances for the anthraquinone moiety, apart from ring A. Resonances at δ 31.1 and 205.7 indicated the presence of an acetyl group, which was positioned at C-12b of ring B by HMBCs from 2-CH3 to the carbonyl carbon C-1 and the quaternary carbon C-12b (δ 142.6). An additional HMBC from 4-CH3 (δ 2.38) to C-4a (δ 139.1) permitted the methyl group to be located at C-4a (Figure 4). At this point, all carbon and proton resonances of 3 were assigned, leading to a structure feature of A-ring cleavage, which is very rare in the angucyclinone/angucycline family. For better comparison, compound 3 was trivially named pratensinon A, and the conventional angucyclinone numbering was used for compounds 1–3 in this paper.
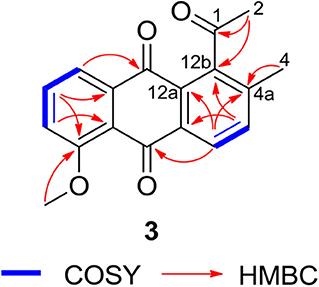
Figure 4. Key 1H-1H COrrelation SpectroscopY (COSY) and heteronuclear multiple bond correlations (HMBCs) of 3.
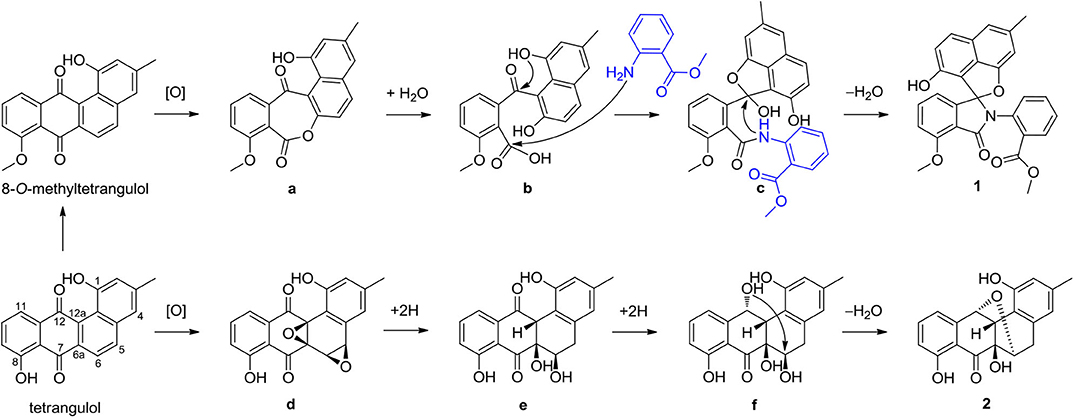
Compound 1 represents a C-ring cleavage angucyclinone with a rare nitrogen-containing spiro ring system that is distinct from the bridged counterpart 2. However, these two compounds might originate from the same angucyclinone precursor tetrangulol (4), which together with 8-O-methyltetrangulol (5), were isolated from the same strain in parallel, and a plausible biosynthetic pathway is proposed in Scheme 1. Starting with methylation on 4 results in ester 5, which on Baeyer–Villiger oxidation gives lactone a, and continues through hydrolytic cleavage to form intermediate b, followed by acetalization and condensation at C-7 ketone by 2-(methoxycarbonyl) aniline to introduce a nitrogen atom in c, which can be intramolecularly cyclized into pratensilin D. In another pathway, oxidation on the double bond Δ5,6 and Δ6a,12a furnishes epoxide d, which undergoes a series of reduction to yield intermediate f, and subsequent cyclization gives rise to kiamycin E.
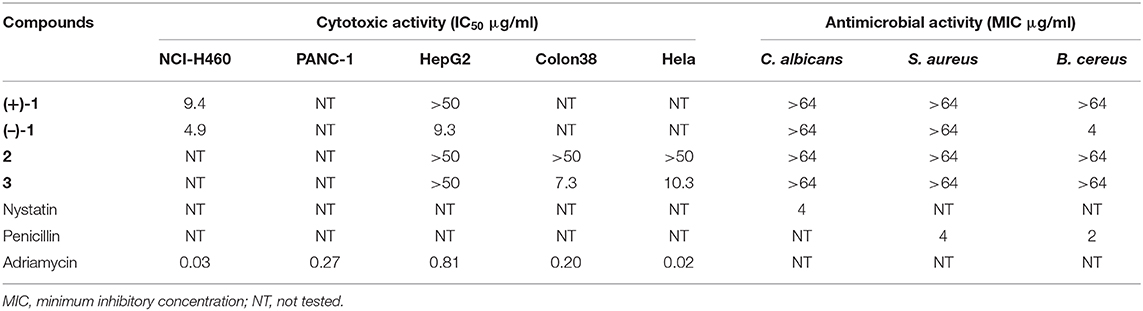
Angucyclines and angucyclinones possess not only great structural diversity but also a broad range of biological activities, predominantly anticancer and antibacterial activities (Kharel et al., 2012). Compounds 1–3 were thus screened for activity against Gram-positive (Bacillus cereus CMCC 32210, Staphylococcus aureus CMCC 26003) bacteria and yeast (Candida albicans CMCC 98001), as well as five human cancer cell lines (NCI-H460, PANC-1, Colon 38, Hela, and HepG2) (Table 2). (–)-1 exhibited selective inhibitory activity against B. cereus CMCC 32210 with a minimum inhibitory concentration (MIC) value of 4 μg/ml; in contrast, its enantiomer (+)-1 showed no efficacy against all tested strains up to 64 μg/ml, so did 2 and 3. Furthermore, (–)-1 exhibited moderate cytotoxicity to NCI-H460 and HepG2 cell lines with respective IC50 values of 4.6 and 9.3 μg/ml, while (+)-1 was only less active to NCI-H460 cells with an IC50 value of 9.2 μg/ml. 3 displayed cytotoxic activity to Colon 38 and Hela cells, with IC50 values of 7.3 and 10.3 μg/ml, respectively. However, 2 showed no inhibitory effect against all tested cell lines in the testing concentration range (IC50 >50 μg/ml).
In summary, five angucyclinones, including three new compounds (1–3) were isolated from the ISP2 medium fermentation with addition of 50 μM LaCl3 of the marine-derived Streptomyces pratensis strain KCB-132. The new compounds possessed different types of structural properties including oxygen bridge (2) and C-ring and A-ring cleavage (1 and 3), respectively. (+)- and (–)-pratensilin D (1), a pair of enantiomeric angucyclinone N-heterocycles, were further separated by chiral HPLC and showed different strengths of biological activities. The new structural properties, especially A-ring and C-ring cleavage disrupting the characteristic tetracyclic ring frame, expand the structural diversity of angucyclines and angucyclinones and provide insight into their structure–activity relationship.
Experimental Section
General Experimental Procedures
Opitical rotations were measured on an Autopol VI (Serial #91058) manufactured by Rudolph Research Analytical, Hackettstown, NJ, USA. UV spectra were recorded by a Shimadzu UV-2401PC spectrometer. CD spectra were recorded using a JASCO J-810 spectropolarimeter. NMR spectra were recorded on Bruker Avance III 600 spectrometers. Electrospray ionization (ESI)-high-resolution mass spectrometry (HRMS) were recorded on an Agilent G6230 TOF spectrometer. Single crystal X-ray crystallography was determined on SMART APEX II DUO X-ray single crystal diffractometer using Cu Kα radiation. Preparative HPLC was performed on a Waters 2489 series instrument with a UV/Visible detector, using a reversed-phase C18 column (Phenomenex, 250 mm × 21.2 mm, 5 μm). Chiral HPLC was carried out on an Agilent 1260 liquid chromatograph, utilizing chiral analytical columns [(R, R) WHELK-01 column, 4.6 mm × 250 mm, 10 μm, 100 A].
Cultivation and Culture Extraction
Streptomyces sp. strain KCB-132 was isolated from a sediment sample collected off Kiaochow Bay, China, as described previously (Zhang et al., 2017). The sequence is deposited in GenBank under accession no. KX033803. The strain KCB-132 was cultured in seawater-based ISP2 medium (5 g of malt extract, 4 g of yeast extract, 4 g of glucose, 500 ml of deionized water, and 500 ml of seawater, pH 7.8) with the addition of 50 μM lanthanum chloride, at a total volume of 21.6 L (72 × 0.3 L), for 10 days at 28°C. The culture broth was filtered to provide filtrate and mycelium. The filtrate was absorbed onto XAD-16 amberlite resin, and the resin was eluted with methanol, then dried out methanol under reduced pressure. The resulting aqueous layer was extracted with ethyl acetate, while the mycelium was extracted by ethyl acetate under ultrasonic radiation directly; both ethyl acetate phases were combined to yield 7.2 g crude extract.
Isolation of Compounds 1–5
The extract (7.2 g) was fractioned by silica gel column chromatography (40 g) and eluted with a step gradient of CH2Cl2 and MeOH. The CH2Cl2/MeOH 99:1 fraction was purified by preparative HPLC (Gemini, C18, 21.2 mm × 250 mm, 5 μm, UV = 210 nm), eluting with 60% MeOH in H2O to afford pratensilin D (1, 1.6 mg), tetrangulol (4, 4.1 mg), and 8-O-methyltetrangulol (5, 3.6 mg). The CH2Cl2/MeOH 49:1 fraction was purified by the same preparative HPLC system eluting with 45% MeOH in H2O to give kiamycin E (2, 1.2 mg) and pratensinon A (3, 0.6 mg). Chiral resolution of 1 was performed on Agilent analytical HPLC system [(R, R) WHELK-01 column, 4.6 mm × 250 mm, 10 μm, 100 A, iso-Pro-OH /n-Hexane = 20:80, 1.0 ml/min, UV = 210 nm] to obtain optically pure (+)-1 (0.6 mg) and (–)-1 (0.6 mg).
(+)-Pratensilin D [(+)-1]: colorless solid; + 50.5 (MeOH, c 0.2); UV (acetonitrile) λmax (log ε) 211 (3.8), 241 (3.5), 302 (2.8), 343 (2.3) nm; 1H-NMR (600 MHz, DMSO-d6) and 13C-NMR (150 MHz, DMSO-d6) data, see Table S1; HRESIMS [M + H]+, m/z 468.14418 (calcd for C28H22O6N, 468.14471).
(–)-Pratensilin D [(–)-1]: colorless solid; – 61.8 (MeOH, c 0.2); UV (acetonitrile) λmax (log ε) 211 (3.9), 241 (3.5), 302 (2.9), 345 (2.4) nm; 1H-NMR (600 MHz, DMSO-d6) and 13C-NMR (150 MHz, DMSO-d6) data, see Table S1; HRESIMS [M + H]+, m/z 468.14418 (calcd for C28H22O6N, 468.14471).
Kiamycin E (2): colorless solid; + 38.7 (MeOH, c 0.2); UV (acetonitrile) λmax (log ε) 203 (3.6), 262 (2.8), 336 (2.5) nm; 1H-NMR (600 MHz, DMSO-d6) and 13C-NMR (150 MHz, DMSO-d6) data, see Table S2; HRESIMS [M + H]+, m/z 325.10740 (calcd for C19H17O5, 325.10760).
Pratensinon A (3): colorless solid; UV (acetonitrile) λmax (log ε) 190 (3.2), 215 (3.09), 257 (3.12), 282 (2.6) nm; 1H-NMR (600 MHz, CDCl3) and 13C-NMR (150 MHz, CDCl3) data, see Table S3; HRESIMS [M + H]+, m/z 295.09677 (calcd for C18H15O4, 295.09703).
X-Ray Crystallographic Analysis
Crystallographic data of 1: X-ray quality crystals were acquired by slow volatilization of a solvent mixture of MeOH and CH2Cl2. Crystal data for xzp6: C28H21NO6, M = 467.46, a = 29.4142(7) Å, b = 12.1834(3) Å, c = 13.7172(3) Å, α = 90°, β = 114.8000(10)°, γ = 90°, V = 4462.42(18) Å3, T = 100. (2) K, space group C12/c1, Z = 8, μ(Cu Kα) = 0.812 mm−1, 41,600 reflections measured, 4,411 independent reflections (Rint = 0.0440). The final R1 values were 0.0419 [I > 2σ(I)]. The final wR(F2) values were 0.1594 [I > 2σ(I)]. The final R1 values were 0.0446 (all data). The final wR(F2) values were 0.1639 (all data). The goodness of fit on F2 was 1.450. Crystallographic data for compound 1 has been deposited in the Cambridge Crystallographic Data Centre with deposition number 1953675.
Crystallographic data of 2: X-ray quality crystals were acquired by slow volatilization of a solvent mixture of MeOH and CH2Cl2. Crystal data for xzp8: C19H16O5, M = 324.32, a = 24.9006(5) Å, b = 8.1765(2) Å, c = 7.4089(2) Å, α = 90°, β = 100.7240(10)°, γ = 90°, V = 1482.11(6) Å3, T = 100. (2) K, space group C121, Z = 4, μ(Cu Kα) = 0.874 mm−1, 13,063 reflections measured, 2,919 independent reflections (Rint = 0.0315). The final R1 values were 0.0281 [I > 2σ(I)]. The final wR(F2) values were 0.0759 [I > 2σ(I)]. The final R1 values were 0.0287 (all data). The final wR(F2) values were 0.0766 (all data). The goodness of fit on F2 was 1.023. Flack parameter = 0.07(5). Crystallographic data for compound 2 has been deposited in the Cambridge Crystallographic Data Centre with deposition number 1992536.
Bioactivity Assay
The antimicrobial assays of compounds 1–3 were tested against Gram-positive (B. cereus CMCC 32210, S. aureus CMCC 26003) bacteria and yeast (C. albicans CMCC 98001) using a microplate assay (Pierce et al., 2008). Nystatin and penicillin were used as positive controls against fungi and bacteria, respectively.
The NCI-H460 (non-small-cell lung carcinoma), PANC-1 (pancreatic cancer), HepG2 (liver hepatocellular carcinoma), Colon 38 (colon cancer), and Hela (cervical carcinoma) cells were plated at a density of 5,000 cells/well in 100 μl Dulbecco's modified Eagle's medium (DMEM). All cell lines were incubated overnight then treated with various concentrations of purified compounds in triplicate. After culturing for 72 h, 20 μl/well of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) solution (5 mg/ml, Sigma-Aldrich, USA) was added to each well, plate was cultured for 4 h at 37°C in a 5% CO2 atmosphere, which was followed by adding 150 μl DMSO to dissolve the formazan crystals, and shaking for 5 min. The absorbance was recorded at 570 nm by a microplate reader. IC50 value was taken using GraphPad Prism 5 software. Adriamycin was used as a positive control.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
LG, LZ, and QY separated and purified the compounds. ML and ZL identified the structures. BX and XF tested cytotoxic and antimicrobial activities. SZ and ZX conceived and designed the experiments. ZX prepared the paper. All authors approved the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Natural Science Foundation of Shandong Province, grant number ZR2018LD006.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2020.00586/full#supplementary-material
References
Arens, J. C., Berru?, F., Pearson, J. K., and Kerr, R. G. (2013). Isolation and structure elucidation of satosporin A and B: new polyketides from Kitasatospora griseola. Org. Lett. 15, 3864–3867. doi: 10.1021/ol401598f
Bethany, K. O., and Mohammad, R. S. (2017). Antibiotic dialogues: induction of silent biosyntheticgene clusters by exogenous small molecules. FEMS Microbiol. Rev. 41, 19–33. doi: 10.1093/femsre/fuw035
Demain, A. L., and Sanchez, S. (2009). Microbial drug discovery: 80 years of progress. J. Antibiot. 62, 5–16. doi: 10.1038/ja.2008.16
Helge, B. B., Barbara, B., Regina, H., and Axel, Z. (2002). Big effects from small changes: possible ways to explore nature's chemical diversity. ChemBioChem. 3, 619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9
Kawai, K., Wang, G., Okamoto, S., and Ochi, K. (2007). The rare earth, scandium, causes antibiotic overproduction in Streptomyces spp. FEMS Microbiol. Lett. 274, 311–315. doi: 10.1111/j.1574-6968.2007.00846.x
Kharel, M. K., Pahari, P., Shepherd, M. D., Tibrewal, N., Nybo, S. E., Shaaban, K. A., et al. (2012). Angucyclines: biosynthesis, mode-of-action, new natural products, and synthesis. J. Nat. Prod. Rep. 29, 264–325. doi: 10.1039/C1NP00068C
Krohn, K., and Rohr, J. (1997). Angucyclines: total syntheses, new structures, and biosynthetic studies of an emerging new class of antibiotics. Top. Curr. Chem. 188, 127–195. doi: 10.1007/BFb0119236
Liu, T., Jin, J., Yang, X., Song, J., Yu, J., Geng, T., et al. (2019). Discovery of a phenylamine-incorporated angucyclinone from marine Streptomyces sp. PKU-MA00218 and generation of derivatives with phenylamine analogues. Org. Lett. 21, 2813–2817. doi: 10.1021/acs.orglett.9b00800
Ma, M., Rateb, M. E., Teng, Q., Yang, D., Rudolf, J. D., Zhu, X., et al. (2015). Angucyclines and angucyclinones from Streptomyces sp. CB01913 featuring C-ring cleavage and expansion. J. Nat. Prod. 78, 2471–2480. doi: 10.1021/acs.jnatprod.5b00601
Newman, D. J., and Cragg, G. M. (2012). Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 75, 311–335. doi: 10.1021/np200906s
Pierce, C. G., Uppuluri, P., Tristan, A. R., Wormley, F. L. Jr., Mowat, E, Ramage, G., et al. (2008). A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat. Protoc. 3, 1494–1500. doi: 10.1038/nprot.2008.141
Rohr, J., and Thiericke, R. (1992). Angucycline group antibiotics. Nat. Prod. Rep. 9, 103–137. doi: 10.1039/np9920900103
Shaaban, K. A., Ahmed, T. A., Leggas, M., and Rohr, J. (2012). Saquayamycins G–K, cytotoxic angucyclines from Streptomyces sp. including two analogues bearing the amino-sugar rednose. J. Nat. Prod. 75, 1383–1392. doi: 10.1021/np300316b
Wu, C., van der Heu, H. U., Melnik, A. V., Lübben, J., Dorrestein, P. C., Minnaard, A. J., et al. (2019). Lugdunomycin, an angucycline-derived molecule with unprecedented chemical architecture. Angew. Chem. Int. Ed. Engl. 58, 2809–2814. doi: 10.1002/anie.201814581
Xie, Z., Zhou, L., Guo, L., Yang, X., Qu, G., Wu, C., et al. (2016). Grisemycin, a bridged angucyclinone with a methylsulfinyl moiety from a marine-derived Streptomyces sp. Org. Lett. 18, 1402–1405. doi: 10.1021/acs.orglett.6b00332
Yixizhuoma Ishikawa, N., Abdelfattah, M. S., and Ishibashi, M. (2017). Elmenols C-H, new angucycline derivatives isolated from a culture of Streptomyces sp. IFM 11490. J. Antibiot. 70, 601–606. doi: 10.1038/ja.2016.158
Keywords: angucyclinones, oxygen bridge, ring cleavage, structure elucidation, Streptomyces pratensis, antibacterial activity, cytotoxicity
Citation: Guo L, Zhang L, Yang Q, Xu B, Fu X, Liu M, Li Z, Zhang S and Xie Z (2020) Antibacterial and Cytotoxic Bridged and Ring Cleavage Angucyclinones From a Marine Streptomyces sp. Front. Chem. 8:586. doi: 10.3389/fchem.2020.00586
Received: 26 March 2020; Accepted: 08 June 2020;
Published: 04 August 2020.
Edited by:
Prasat Kittakoop, Chulabhorn Graduate Institute, ThailandReviewed by:
Yongbo Xue, Huazhong University of Science and Technology, ChinaHou-Wen Lin, Shanghai Jiao Tong University, China
Copyright © 2020 Guo, Zhang, Yang, Xu, Fu, Liu, Li, Zhang and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zeping Xie, zepingxie@sina.com; Shumin Zhang, shumin_zhang@outlook.com
 Lin Guo1
Lin Guo1  Zeping Xie
Zeping Xie