Abstract
Autophagy supports both cellular and organismal homeostasis. However, whether autophagy should be inhibited or activated for cancer therapy remains unclear. Deletion of essential autophagy genes increased the sensitivity of mouse mammary carcinoma cells to radiation therapy in vitro and in vivo (in immunocompetent syngeneic hosts). Autophagy-deficient cells secreted increased amounts of type I interferon (IFN), which could be limited by CGAS or STING knockdown, mitochondrial DNA depletion or mitochondrial outer membrane permeabilization blockage via BCL2 overexpression or BAX deletion. In vivo, irradiated autophagy-incompetent mammary tumors elicited robust immunity, leading to improved control of distant nonirradiated lesions via systemic type I IFN signaling. Finally, a genetic signature of autophagy had negative prognostic value in patients with breast cancer, inversely correlating with mitochondrial abundance, type I IFN signaling and effector immunity. As clinically useful autophagy inhibitors are elusive, our findings suggest that mitochondrial outer membrane permeabilization may represent a valid target for boosting radiation therapy immunogenicity in patients with breast cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. The METABRIC patient dataset can be publicly accessed via cBioPortal at https://www.cbioportal.org/study/summary?id=brca_metabric. The Molecular Signature Database is publicly available at https://www.gsea-msigdb.org/gsea/msigdb/index.jsp. Source data are provided with this paper.
Code availability
The code employed for in silico studies has been deposited at GitHub and is publicly available at https://github.com/icbi-lab/Yamazaki_et_al_Nature_Immunology_2020. Source data are provided with this paper.
References
Galluzzi, L. et al. Molecular definitions of autophagy and related processes. EMBO J. 36, 1811–1836 (2017).
Dikic, I. & Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19, 349–364 (2018).
Galluzzi, L., Bravo-San Pedro, J. M., Levine, B., Green, D. R. & Kroemer, G. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 16, 487–511 (2017).
Michaud, M. et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science 334, 1573–1577 (2011).
Levy, J. M. M., Towers, C. G. & Thorburn, A. Targeting autophagy in cancer. Nat. Rev. Cancer 17, 528–542 (2017).
Rybstein, M. D., Bravo-San Pedro, J. M., Kroemer, G. & Galluzzi, L. The autophagic network and cancer. Nat. Cell Biol. 20, 243–251 (2018).
Galluzzi, L., Chan, T. A., Kroemer, G., Wolchok, J. D. & Lopez-Soto, A. The hallmarks of successful anticancer immunotherapy. Sci. Transl. Med. 10, eaat7807 (2018).
Ngwa, W. et al. Using immunotherapy to boost the abscopal effect. Nat. Rev. Cancer 18, 313–322 (2018).
Rodriguez-Ruiz, M. E., Vitale, I., Harrington, K. J., Melero, I. & Galluzzi, L. Immunological impact of cell death signaling driven by radiation on the tumor microenvironment. Nat. Immunol. 21, 120–134 (2020).
Formenti, S. C. et al. Radiotherapy induces responses of lung cancer to CTLA-4 blockade. Nat. Med. 24, 1845–1851 (2018).
Demaria, S. et al. Ionizing radiation inhibition of distant untreated tumors (abscopal effect) is immune mediated. Int. J. Radiat. Oncol. Biol. Phys. 58, 862–870 (2004).
Dewan, M. Z. et al. Fractionated but not single-dose radiotherapy induces an immune-mediated abscopal effect when combined with anti-CTLA-4 antibody. Clin. Cancer Res. 15, 5379–5388 (2009).
Vanpouille-Box, C. et al. DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nat. Commun. 8, 15618 (2017).
Mackenzie, K. J. et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017).
Harding, S. M. et al. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 (2017).
Sprooten, J., Agostinis, P. & Garg, A. D. Type I interferons and dendritic cells in cancer immunotherapy. Int. Rev. Cell Mol. Biol. 348, 217–262 (2019).
Zierhut, C. et al. The cytoplasmic DNA sensor cGAS promotes mitotic cell death. Cell 178, 302–315 (2019).
Abe, T. & Shapira, S. D. Negative regulation of cytosolic sensing of DNA. Int. Rev. Cell Mol. Biol. 344, 91–115 (2019).
Singh, R., Letai, A. & Sarosiek, K. Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 20, 175–193 (2019).
McArthur, K. et al. BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science 359, eaao6047 (2018).
Sliter, D. A. et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature 561, 258–262 (2018).
Lindqvist, L. M. et al. Autophagy induced during apoptosis degrades mitochondria and inhibits type I interferon secretion. Cell Death Differ. 25, 784–796 (2018).
Rongvaux, A. et al. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell 159, 1563–1577 (2014).
White, M. J. et al. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell 159, 1549–1562 (2014).
Rodriguez-Ruiz, M. E. et al. Apoptotic caspases inhibit abscopal responses to radiation and identify a new prognostic biomarker for breast cancer patients. Oncoimmunology 8, e1655964 (2019).
Galluzzi, L., Bravo-San Pedro, J. M., Demaria, S., Formenti, S. C. & Kroemer, G. Activating autophagy to potentiate immunogenic chemotherapy and radiation therapy. Nat. Rev. Clin. Oncol. 14, 247–258 (2017).
Golden, E. B. et al. Radiation fosters dose-dependent and chemotherapy-induced immunogenic cell death. Oncoimmunology 3, e28518 (2014).
Galluzzi, L. et al. Consensus guidelines for the definition, detection and interpretation of immunogenic cell death. J. Immunother. Cancer 8, e000337 (2020).
Galluzzi, L., Buque, A., Kepp, O., Zitvogel, L. & Kroemer, G. Immunogenic cell death in cancer and infectious disease. Nat. Rev. Immunol. 17, 97–111 (2017).
Bartsch, K. et al. Absence of RNase H2 triggers generation of immunogenic micronuclei removed by autophagy. Hum. Mol. Genet. 26, 3960–3972 (2017).
Walczak, J., Partyka, M., Duszynski, J. & Szczepanowska, J. Implications of mitochondrial network organization in mitochondrial stress signalling in NARP cybrid and Rho0 cells. Sci. Rep. 7, 14864 (2017).
Galluzzi, L. et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 25, 486–541 (2018).
Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012).
Vitale, I., Galluzzi, L., Castedo, M. & Kroemer, G. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat. Rev. Mol. Cell Biol. 12, 385–392 (2011).
Ning, X. et al. Apoptotic caspases suppress type I interferon production via the cleavage of cGAS, MAVS, and IRF3. Mol. Cell 74, e17 (2019).
Han, C. et al. Tumor cells suppress radiation-induced immunity by hijacking caspase 9 signaling. Nat. Immunol. 21, 546–554 (2020).
Luo, S. & Rubinsztein, D. C. Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ. 17, 268–277 (2010).
Fairlie, W. D., Tran, S. & Lee, E. F. Crosstalk between apoptosis and autophagy signaling pathways. Int Rev. Cell Mol. Biol. 352, 115–158 (2020).
Rodriguez-Ruiz, M. E. et al. Abscopal effects of radiotherapy are enhanced by combined immunostimulatory mAbs and are dependent on CD8 T cells and crosspriming. Cancer Res. 76, 5994–6005 (2016).
Vanpouille-Box, C. et al. TGFβ is a master regulator of radiation therapy-induced antitumor immunity. Cancer Res. 75, 2232–2242 (2015).
Jain, N. et al. Ibrutinib and venetoclax for first-line treatment of CLL. N. Engl. J. Med. 380, 2095–2103 (2019).
Lok, S. W. et al. A phase Ib dose-escalation and expansion study of the BCL2 inhibitor venetoclax combined with tamoxifen in ER and BCL2-positive metastatic breast cancer. Cancer Discov. 9, 354–369 (2019).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Galluzzi, L. et al. Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ. 16, 1093–1107 (2009).
Morselli, E. et al. p53 inhibits autophagy by interacting with the human ortholog of yeast Atg17, RB1CC1/FIP200. Cell Cycle 10, 2763–2769 (2011).
Michels, J. et al. Cisplatin resistance associated with PARP hyperactivation. Cancer Res. 73, 2271–2280 (2013).
McQuin, C. et al. CellProfiler 3.0: next-generation image processing for biology. PLoS Biol. 16, e2005970 (2018).
Fan, A. X. et al. Mitochondrial DNA content in paired normal and cancerous breast tissue samples from patients with breast cancer. J. Cancer Res. Clin. Oncol. 135, 983–989 (2009).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000).
The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338 (2019).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011).
Mootha, V. K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Acknowledgements
The authors are indebted to B. Vogelstein (Johns Hopkins University) for sharing BAX+/− and BAX−/− HCT 116 cells and the WCM Radiation Biology Core Facility (in particular J. Kraynak) for help with irradiation procedures. F.F. is supported by the Austrian Science Fund (FWF) (project no. T 974-B30). E.N.M. is supported by an FPU fellowship from Ministerio de Educación Cultura y Deporte of Spain (FPU16/06537). S.C.F. is supported by Breakthrough Level 2 and 3 grants from the US Department of Defense Breast Cancer Research Program (BC180476, PI: Formenti; BC180595, PI: Formenti, Demaria) and by an S10 Instrumentation Grant from National Institutes of Health (RR027619-01). The L.G. laboratory is supported by a Breakthrough Level 2 grant from the US Department of Defense Breast Cancer Research Program (BC180476P1, PI: Galluzzi), by the 2019 Laura Ziskin Prize in Translational Research (ZP-6177, PI: Formenti) from Stand Up to Cancer, by a Mantle Cell Lymphoma Research Initiative (PI: Chen-Kiang) grant from the Leukemia and Lymphoma Society, by a startup grant from the Department of Radiation Oncology at Weill Cornell Medicine, by a Rapid Response Grant from the Functional Genomics Initiative, by industrial collaborations with Lytix Biopharma and Phosplatin Therapeutics and by donations from Phosplatin, the Luke Heller TECPR2 Foundation and Sotio a.s.
Author information
Authors and Affiliations
Contributions
T.Y. performed the majority of experimental assessments with the help of A.S. and M.R. and prepared figures with the help of A.B. and NB. A.B., G.P., N.B. and L.S. provided additional experimental support. A.B. performed automated image analyses. A.K. performed in silico analyses with support from F.F. and under supervision by Z.T. E.N.M. performed qPCR on patient samples. F.A.P. and E.G.M. identified patient samples and performed statistical analyses on patient data with support from A.K. S.C.F. provided infrastructure and clinical input on the project. L.G. conceived the project, directed experimental and biostatistical assessments, interpreted data, wrote the manuscript and supervised figure preparation. All authors approved the final version of the paper.
Corresponding author
Ethics declarations
Competing interests
F.A.P. declares research funding from Celgene and Roche and speaker and/or advisory honoraria from Pfizer, Roche, AstraZeneca, Celgene, Eisai, Novartis, Pierre Fabre and Roche. E.G.M. declares research funding from Roche and speaker and/or advisory honoraria from AstraZeneca, Roche and Pharmamar. S.C.F. declares funding for clinical trials from Bristol Myers Squibb, Merck and Varian and speaker and/or advisory honoraria from Astra Zeneca, Bayer, Bristol Myers Squibb, Eisai, Elekta, EMD Serono/Merck, GlaxoSmithKline, Janssen, MedImmune, Merck US, Regeneron, Varian and ViewRay. L.G. declares research funding from Lytix Biopharma and Phosplatin Therapeutics and speaker and/or advisory honoraria from Boehringer Ingelheim, Astra Zeneca, OmniSEQ, the Longevity Labs, Inzen and the Luke Heller TECPR2 Foundation.
Additional information
Editor recognition statement Zoltan Fehervari was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
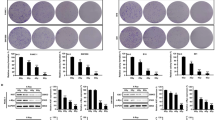
Extended Data Fig. 1 Autophagy supports radioresistance in vitro and in vivo.
a, Protein levels of unconjugated ATG5, ATG5-ATG12 conjugates, and ATG7 in control (SCR) and Atg5–/– or Atg7–/– TS/A cell clones generated for the purpose of this study, as determined by immunoblotting. ACTB levels were monitored to ensure equal lane loading. b, Percentage of TS/A cells preserving plasma membrane integrity, as assessed by the vital dye propidium iodide (PI), 24 h after exposure to the γ irradiation (4 Gy) in the optional presence of 25 µM hydroxychloroquine (HCQ). Results are means ± SEM and individual data points. Number of biologically independent samples collected over 5 (Control, 4 Gy) or two (HCQ, HCQ + 4 Gy) independent experiments and p values (one way-ANOVA plus Fisher LSD as compared to untreated TS/A cells*, unpaired two-sided Student’s t test as compared to irradiated TS/A cells#) are reported. c, Tumor growth (individual curves) and overall survival (OS) of immunocompetent BALB/c mice implanted with SCR, Atg5–/– and Atg7–/– TS/A cells and optionally treated with intraperitoneal mitoxantrone (MTX). Number of animals and p values (two-sided log-rank as compared to untreated mice bearing lesions of the same genotype) are reported. d, Percentage of tumor-free BALB/c mice upon challenge with a tumorigenic number of living SCR TS/A cells 2 weeks days after vaccination with PBS (negative control) or TS/A cells of the indicated genotype pre-cultured for 24 h with 1 µM MTX. Number of animals, tumor-free survival (TFS) at the end of the experiment, and p values (two-sided log-rank as compared to PBS vaccination* or vaccination with irradiated SCR cells#) are reported.
Extended Data Fig. 2 Autophagy inhibits type I IFN secretion by irradiated cancer cells as a consequence of improved cytosolic DNA clearance.
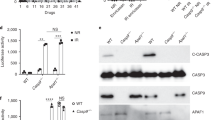
a-d, Ifnb1 (a, c) or secreted type I IFN (b, d) levels in control (SCR) or Atg7–/– mouse mammary carcinoma EO771 cells (a, b), wild-type mouse fibrosarcoma MCA205 cells (c), and wild-type human mammary carcinoma MCF7 cells (d), optionally exposed to γ irradiation (8 Gy) and cultured in control conditions or in the presence of 10–25 µM hydroxychloroquine (HCQ) for 24 h, as assessed by RT-PCR (a, c) or ELISA (b, d). Results are means ± SEM and individual data points. Number of biologically independent samples collected over three (a, c, d) or two (b) independent experiments and p values (one way-ANOVA plus Fisher LSD as compared to untreated SCR cells*, unpaired two-sided Student’s t test as compared to irradiated wild-type or SCR cells#) are reported. n.d., not detectable above blank. ATG7 levels and ATG5 conjugation status as assessed by immunoblotting are depicted. ACTB levels were monitored to ensure equal lane loading. e, Ifnb1 levels in Atg7–/– TS/A cells optionally exposed to γ irradiation (8 Gy) and then cultured in control conditions or in the presence of 10 µM RU320521 (RU.521) for 48 h, as assessed by quantitative RT-PCR. Results are means ± SEM and individual data points. Number of biologically independent samples and p values (paired two-sided Student’s t test as compared to irradiated Atg7–/– cells#) are reported.
Extended Data Fig. 3 Autophagy inhibits type I IFN secretion by limiting cytosolic mtDNA accumulation.
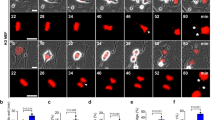
a, Micronucleation in wild-type TS/A cells optionally exposed to γ irradiation at the indicated dose (8 Gy) and maintained in control conditions for 24 h. Representative images (scale bar = 30 µm) and quantitative data are reported. Results are means ± SEM and individual data points. Number of images from biologically independent samples collected over three independent experiments and p value (unpaired two-sided Student’s t test as compared to untreated cells*) are reported. b, Cytosolic dsDNA species in rho0 TS/A cells exposed to γ irradiation at the indicated dose (8 Gy) and maintained in control conditions for 24 h, as assessed by high resolution confocal microscopy upon immunofluorescence staining with antibodies specific for dsDNA and COX4. DAPI was employed as nuclear counterstain. Representative images (scale bar = 20 μm) are reported from a total of 9 (control) and 13 (8 Gy) collected from biologically independent samples over three independent experiments. c. COX4 levels in wild-type and rho0 TS/A cells, as assessed by immunoblotting with COX4-specific antibodies. ACTB levels were assessed to ensure equal lane loading. d, BCL2 levels in wild-type TS/A cells and TS/A cells transiently transfected with a commercial plasmid for BCL2-overexpression (BCL2++), as assessed by immunoblotting with BCL2-specific antibodies. ACTB levels were assessed to ensure equal lane loading. e, f, Amount of cytosolic dsDNA in wild-type and BCL2-overexpressing (BCL2++) TS/A cells (e) or BAX+/– and BAX–/– human colorectal carcinoma HCT 116 cells (f) optionally exposed to γ irradiation (8 Gy, e; 20 Gy, f) and cultured in control conditions for 48 h, as assessed by conventional immunofluorescence microscopy and automated image analysis. DAPI was employed as nuclear counterstain. Representative images (scale bar = 20 μm) from a total of 17 (e, wild-type TS/A cells), 18 (e, BCL2++ TS/A cells), 10 (f, BAX+/– HCT 116 cells) and 18 (f, BAX–/– HCT 116 cells) collected from biologically independent samples over three independent experiments. g. Mitochondrial (mt) and genomic (g) DNA levels in control (SCR), Atg5–/– and Atg7–/– TS/A cells maintained in control conditions or subjected to mtDNA depletion by long-term exposure to ethidium bromide (rho0), as assessed by quantitative PCR with specific primers. Densitometry upon normalization to gDNA is reported.
Extended Data Fig. 4 Impact of autophagic proficiency and mtDNA levels on clinical breast cancer outcome.
a, b, Unsupervised hierarchical clustering of top 400 differentially expressed genes (DEGs) in 1351 breast cancer patients from the METABRIC database for whom cancer-specific overall survival (CSOS) data are available, upon median stratification of a gene signature based on the z-scored median expression of ATG5, ATG7, ATG10, ATG12 and ATG16L1 (ATG signature). Gene set enrichment analysis (GSEA) for the GO terms “response to type I interferon” and “response to interferon gamma”, as well as for the Hallmarks (HL) terms “interferon alpha response” and “interferon gamma response” is depicted. Normalized enrichment scores (NES) and false discovery rate (FDR)-adjusted p values (q values as calculated in the GSEA analysis) are reported. c, GSEA for the GO term “response to gamma radiation” in all 1820 breast cancer patients from the METABRIC database, as well as in 1351 patients for whom CSOS information is available upon median stratification based on the ATG signature. NES and FDR-adjusted p values (q values as calculated in the GSEA analysis) are reported. d, Intensity of a genetic signature representative of mitochondrial abundance (Mitoonly) in the tumor microenvironment of 1820 unselected breast cancer patients from the METABRIC database stratification by median, tertiles or quartiles of the ATG signature. p values (Kruskal Wallis) are reported. Boxplots represent median, upper and lower quartiles, and additional points within 1.5 times the interquartile range from upper or lower quartiles. e, Disease-free survival (DFS) and overall survival (OS) of 37 breast cancer patients from the University Hospital of Murcia for which bioptic material from the primary tumor was available, upon median stratification based on mitochondrial (mt) to genomic (g) DNA ratio. p values (two-sided log-rank) are reported. See also Extended Data Table 3. f, DFS of 9 breast cancer patients from the University Hospital of Murcia for which bioptic material from metastatic lesions at relapse or progressive disease was available, upon median stratification based on mitochondrial (mt) to genomic (g) DNA ratio. p values (two-sided log-rank) are reported.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2.
Supplementary Video 1
Z-stack reconstruction of wild-type TS/A cells maintained in control conditions then stained with DAPI (nuclear counterstain) plus dsDNA-specific (red) and LMNB-specific (green) antibodies. See also Fig. 3a.
Supplementary Video 2
Z-stack reconstruction of wild-type TS/A cells exposed to γ irradiation (8 Gy) and cultured in control conditions for 24 h then stained with DAPI (nuclear counterstain) plus dsDNA-specific (red) and LMNB-specific (green) antibodies. See also Fig. 3a.
Supplementary Video 3
Z-stack reconstruction of wild-type TS/A cells maintained in control then stained with DAPI (nuclear counterstain) plus dsDNA-specific (red) and COX4-specific (green) antibodies. See also Fig. 3c.
Supplementary Video 4
Z-stack reconstruction of wild-type TS/A cells exposed to γ irradiation (8 Gy) and cultured in control conditions for 24 h then stained with DAPI (nuclear counterstain) plus dsDNA-specific (red) and COX4-specific (green) antibodies. See also Fig. 3c.
Supplementary Video 5
Z-stack reconstruction of wild-type TS/A cells maintained in control conditions then stained with DAPI (nuclear counterstain) plus dsDNA-specific (red) and TFAM-specific (green) antibodies. See also Fig. 3e.
Supplementary Video 6
Z-stack reconstruction of wild-type TS/A cells exposed to γ irradiation (8 Gy) and cultured in control conditions for 24 h then stained with DAPI (nuclear counterstain) plus dsDNA-specific (red) and TFAM-specific (green) antibodies. See also Fig. 3e.
Supplementary Video 7
Z-stack reconstruction of mtDNA-depleted TS/A cells maintained in control conditions then stained with DAPI (nuclear counterstain) plus dsDNA-specific (red) and LMNB-specific (green) antibodies. See also Fig. 4c.
Supplementary Video 8
Z-stack reconstruction of mtDNA-depleted TS/A cells exposed to γ irradiation (8 Gy) and cultured in control conditions for 24 h then stained with DAPI (nuclear counterstain) plus dsDNA-specific (red) and LMNB-specific (green) antibodies. See also Fig. 4c.
Supplementary Table 1
Differential gene expression analysis on the METABRIC cohort and survival analysis on the METABRIC and Murcia cohort.
Source data
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 3
Unprocessed gels.
Source Data Fig. 4
Unprocessed gels.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 3
Unprocessed western blots and gels.
Rights and permissions
About this article
Cite this article
Yamazaki, T., Kirchmair, A., Sato, A. et al. Mitochondrial DNA drives abscopal responses to radiation that are inhibited by autophagy. Nat Immunol 21, 1160–1171 (2020). https://doi.org/10.1038/s41590-020-0751-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-020-0751-0
This article is cited by
-
Targeting immunogenic cell stress and death for cancer therapy
Nature Reviews Drug Discovery (2024)
-
Cancer cell metabolism and antitumour immunity
Nature Reviews Immunology (2024)
-
MRE11 mobilizes CGAS and drives ZBP1-dependent necroptosis
Cell Research (2024)
-
Inflammation and mitophagy are mitochondrial checkpoints to aging
Nature Communications (2024)
-
Immunological aspects of central neurodegeneration
Cell Discovery (2024)