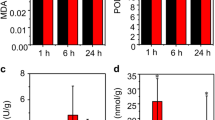
Abstract
Upland cotton (Gossypium hirsutum) is a salt-tolerant crop that can endure high salt concentrations without apparent damage. However, the plant’s response to salinity stress is a complex biological process. An analysis of the dynamic changes in transcript profiles will provide a global picture of the cotton response to salinity stress. Here, we monitored the transcriptome changes in two cotton genotypes, the salt-tolerant H15, and sensitive ZM12, at 0, 0.25, 1, 3, 6, 12, 24, and 48 h in roots exposed to 200-mM NaCl. In total, 13,894 and 5057 differentially expressed genes were identified as being involved in salt-stress tolerance in H15 and ZM12, respectively. Of these, 3825 genes were common to both genotypes. A differential expression analysis revealed that the number of differentially expressed genes increased significantly during the first 24 h after the salt-stress treatment and then significantly decreased at 48 h in both genotypes. A transcription factor (TF) analysis revealed three different patterns based on the expression of 45 TFs’ families, with the majority of differentially expressed TFs increasing rapidly after the salt-stress treatment in both genotypes. A weighted gene co-expression network analysis showed that two gene modules were related to salinity, and genes in these modules were mainly involved in plant–pathogen interactions, the plant MAPK signaling pathway, and diterpenoid biosynthesis. Our results increase the understanding of cotton metabolic pathways involved in responses to salt stress.









Similar content being viewed by others
Abbreviations
- BCAT:
-
Branched-chain aminotransferase
- CAT:
-
Catalase
- CPKs/CDPKs:
-
Calcium-dependent protein kinases
- DEG:
-
Differentially expressed gene
- FPKM:
-
Fragments per kilobase per million
- GO:
-
Gene ontology
- GPX:
-
Glutathione peroxidase
- GR:
-
Glutathione reductase
- HAK:
-
High-affinity K+ transporter
- HMGR:
-
Hydroxy methylglutaryl CoA reductase
- HRGPs:
-
Hydroxyproline-rich glycoproteins
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- MAPK:
-
Mitogen-activated protein kinase
- MYB:
-
Myb domain protein
- PANK:
-
Pantothenate kinase genes
- POD:
-
Peroxidase
- REC:
-
Relative electrical conductivity
- ROS:
-
Reactive oxygen species
- RWC:
-
Relative water content
- TF:
-
Transcription factor
References
Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311(5757):91–94. https://doi.org/10.1126/science.1118642
Alemán F, Nieves-Cordones M, Martínez V, Rubio F (2009) Differential regulation of the HAK5 genes encoding the high-affinity K+ transporters of Thellungiella halophila and Arabidopsis thaliana. Environ Exp Bot 65(2–3):263–269. https://doi.org/10.1016/j.envexpbot.2008.09.011
Almadanim MC, Alexandre BM, Rosa MT, Sapeta H, Leitão AE, Ramalho JC, Lam TT, Negrão S, Abreu IA, Oliveira MM (2017) Rice calcium-dependent protein kinase OsCPK17 targets plasma membrane intrinsic protein and sucrose phosphate synthase and is required for a proper cold stress response. Plant Cell Environ 40(7):1197–1213
Antolín-Llovera M, Leivar P, Arró M, Ferrer A, Boronat A, Campos N (2014) Modulation of plant HMG-CoA reductase by protein phosphatase 2A. Plant Signal Behav 6(8):1127–1131. https://doi.org/10.4161/psb.6.8.16363
Asano T, Hayashi N, Kikuchi S, Ohsugi R (2012) CDPK-mediated abiotic stress signaling. Plant Signal Behav 7(7):817–821
Batelli G, Verslues PE, Agius F, Qiu Q, Fujii H, Pan S, Schumaker KS, Grillo S, Zhu J (2007) SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Mol Cell Biol 27(22):7781–7790. https://doi.org/10.1128/MCB.00430-07
Bibi Z, Khan NU, Latif A, Khan MJ, Manyuan G, Niu Y, Khan QU, Khan IU, Shaheen S, Sadozai GU (2016) Salt tolerance in upland cotton genotypes to nacl salinity at early growth stages. J Anim Plant Sci 26(3):766
Brinker M, Brosché M, Vinocur B, Abo-Ogiala A, Fayyaz P, Janz D, Ottow EA, Cullmann AD, Saborowski J, Kangasjärvi J, Altman A, Polle A (2010) Linking the salt transcriptome with physiological responses of a salt-resistant populus species as a strategy to identify genes important for stress acclimation. Plant Physiol 154(4):1697–1709. https://doi.org/10.1104/pp.110.164152
Chen Z, Xie Y, Gu Q, Zhao G, Zhang Y, Cui W, Xu S, Wang R, Shen W (2017) The AtrbohF -dependent regulation of ROS signaling is required for melatonin-induced salinity tolerance in Arabidopsis. Free Radic Biol Med 108:465–477. https://doi.org/10.1016/j.freeradbiomed.2017.04.009
Chen C, Xia R, Chen H, He Y (2018a) TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv. https://doi.org/10.1101/289660
Chen Y, Ren Y, Zhang G, An J, Yang J, Wang Y, Wang W (2018b) Overexpression of the wheat expansin gene TaEXPA2 improves oxidative stress tolerance in transgenic Arabidopsis plants. Plant Physiol Biochem 124:190–198. https://doi.org/10.1016/j.plaphy.2018.01.020
Choi HI, Kim YW, Hwang I, Kim SY (2005) Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiol 139(4):1750–1761
Chu X, Wang C, Chen X, Lu W, Li H, Wang X, Hao L, Guo X (2016) Correction: the cotton WRKY gene GhWRKY41 positively regulates salt and drought stress tolerance in transgenic Nicotiana benthamiana. PLoS One 10(11):e143022
Çiçek N, Çakirlar H (2008) Changes in some antioxidant enzyme activities in six soybean cultivars in response to long-term salinity at two different temperatures. Gen Appl Plant Physiol 34:267–280
Diaz-Lopez L, Gimeno V, Lidon V, Simon I, Martinez V, Garcia-Sanchez F (2012) The tolerance of Jatropha curcas seedlings to NaCl: an ecophysiological analysis. Plant Physiol Biochem 54:34–42. https://doi.org/10.1016/j.plaphy.2012.02.005
Ding Z, Li S, An X, Liu X, Qin H, Wang D (2009) Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J Genet Genomics 36(1):17–29. https://doi.org/10.1016/S1673-8527(09)60003-5
Droillard MJ, Boudsocq M, Barbier-Brygoo H, Laurière C (2002) Different protein kinase families are activated by osmotic stresses in Arabidopsis thaliana cell suspensions: involvement of the MAP kinases AtMPK3 and AtMPK6. FEBS Lett 527(1):43–50
Duan F, Ding J, Lee D, Lu X, Feng Y, Song W (2017) Overexpression of SoCYP85A1, a spinach cytochrome p450 gene in transgenic tobacco enhances root development and drought stress tolerance. Front Plant Sci. https://doi.org/10.3389/fpls.2017.01909
Gao SQ, Chen M, Xia LQ, Xiu HJ, Xu ZS, Li LC, Zhao CP, Cheng XG, Ma YZ (2009) A cotton (Gossypium hirsutum) DRE-binding transcription factor gene, GhDREB, confers enhanced tolerance to drought, high salt, and freezing stresses in transgenic wheat. Plant Cell Rep 28(2):301–311. https://doi.org/10.1007/s00299-008-0623-9
Guengerich FP (2007) Mechanisms of cytochrome P450 substrate oxidation: MiniReview. J Biochem Mol Toxicol 21(4):163–168
Guo YH, Yu YP, Wang D, Wu CA, Yang GD, Huang JG, Zheng CC (2009) GhZFP1, a novel CCCH-type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5. New Phytol 183(1):62–75. https://doi.org/10.1111/j.1469-8137.2009.02838.x
Guo J, Shi G, Guo X, Zhang L, Xu W, Wang Y, Su Z, Hua J (2015) Transcriptome analysis reveals that distinct metabolic pathways operate in salt-tolerant and salt-sensitive upland cotton varieties subjected to salinity stress. Plant Sci 238:33–45
Gupta B, Huang B (2014) Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int J Genomics 2014:701596. https://doi.org/10.1155/2014/701596
Haake V (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130(2):639–648. https://doi.org/10.1104/pp.006478
Hamamoto S, Horie T, Hauser F, Deinlein U, Schroeder JI, Uozumi N (2015) HKT transporters mediate salt stress resistance in plants: from structure and function to the field. Curr Opin Biotechnol 32:113–120. https://doi.org/10.1016/j.copbio.2014.11.025
Huang GQ, Li W, Zhou W, Zhang JM, Li DD, Gong SY, Li XB (2013) Seven cotton genes encoding putative NAC domain proteins are preferentially expressed in roots and in responses to abiotic stress during root development. Plant Growth Regul 71(2):101–112
Huang J, Duan XW, Zhang WN, Meng D, Chao MA, Hao L, Tian-Zhong LI (2015) Clone of pear HMGRgene and salt-tolerance analysis of its transgenic tobacco seed. J Chin Agric Univ 20(1):60–67
Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K (2000) Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J 24(5):655–665. https://doi.org/10.1046/j.1365-313x.2000.00913.x
Jin J, Tian F, Yang D, Meng Y, Kong L, Luo J, Gao G (2017) PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res 45(D1):D1040–D1045. https://doi.org/10.1093/nar/gkw982
Karve RA (2009) Functional characterization of calcium dependent protein kinase 32 from Arabidopsis. Dissertations and Theses - Gradworks
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12(4):357–360. https://doi.org/10.1038/nmeth.3317
Kline KG, Barrett-Wilt GA, Sussman MR (2010) In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc Natl Acad Sci 107(36):15986–15991. https://doi.org/10.1073/pnas.1007879107
Konig S, Mosblech A, Heilmann I (2007) Stress-inducible and constitutive phosphoinositide pools have distinctive fatty acid patterns in Arabidopsis thaliana. FASEB J 21(9):1958–1967. https://doi.org/10.1096/fj.06-7887com
Kotchoni SO, Kuhns C, Ditzer A, Kirch H, Bartels D (2006) Over-expression of different aldehyde dehydrogenase genes in Arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant Cell Environ 29(6):1033–1048. https://doi.org/10.1111/j.1365-3040.2005.01458.x
Kronzucker HJ, Britto DT (2011) Sodium transport in plants: a critical review. New Phytol 189(1):54–81. https://doi.org/10.1111/j.1469-8137.2010.03540.x
Kurotani K, Hayashi K, Hatanaka S, Toda Y, Ogawa D, Ichikawa H, Ishimaru Y, Tashita R, Suzuki T, Ueda M, Hattori T, Takeda S (2015) Elevated levels of CYP94 family gene expression alleviate the jasmonate response and enhance salt tolerance in rice. Plant Cell Physiol 56(4):779–789. https://doi.org/10.1093/pcp/pcv006
Kwon YR, Lee HJ, Kim KH, Hong SW, Lee SJ, Lee H (2008) Ectopic expression of Expansin3 or Expansinbeta1 causes enhanced hormone and salt stress sensitivity in Arabidopsis. Biotechnol Lett 30(7):1281–1288. https://doi.org/10.1007/s10529-008-9678-5
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinform 9:559. https://doi.org/10.1186/1471-2105-9-559
Li F, Fan G, Lu C, Xiao G, Zou C, Kohel RJ, Ma Z, Shang H, Ma X, Wu J, Liang X, Huang G, Percy RG, Liu K, Yang W, Chen W, Du X, Shi C, Yuan Y, Ye W, Liu X, Zhang X, Liu W, Wei H, Wei S, Huang G, Zhang X, Zhu S, Zhang H, Sun F, Wang X, Liang J, Wang J, He Q, Huang L, Wang J, Cui J, Song G, Wang K, Xu X, Yu JZ, Zhu Y, Yu S (2015a) Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol 33(5):524–530. https://doi.org/10.1038/nbt.3208
Li W, Zhao F, Fang W, Xie D, Hou J, Yang X, Zhao Y, Tang Z, Nie L, Lv S (2015b) Identification of early salt stress responsive proteins in seedling roots of upland cotton (Gossypium hirsutum L.) employing iTRAQ-based proteomic technique. Front Plant Sci 6:732. https://doi.org/10.3389/fpls.2015.00732
Liu Y, He C (2017) A review of redox signaling and the control of MAP kinase pathway in plants. Redox Biol 11(3):192–204. https://doi.org/10.1016/j.redox.2016.12.009
Lo SF, Ho THD, Liu YL, Jiang MJ, Hsieh KT, Chen KT, Yu LC, Lee MH, Chen CY, Huang TP, Kojima M, Sakakibara H, Chen LJ, Yu SM (2017) Ectopic expression of specific GA2 oxidase mutants promotes yield and stress tolerance in rice. Plant Biotechnol J 15(7):850–864. https://doi.org/10.1111/pbi.12681
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550. https://doi.org/10.1186/s13059-014-0550-8
Ma S, Bohnert HJ (2007) Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biol 8(4):R49. https://doi.org/10.1186/gb-2007-8-4-r49
Malatrasi M, Corradi M, Svensson JT, Close TJ, Gulli M, Marmiroli N (2006) A branched-chain amino acid aminotransferase gene isolated from Hordeum vulgare is differentially regulated by drought stress. Theor Appl Genet 113(6):965–976. https://doi.org/10.1007/s00122-006-0339-6
Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA, Okamoto M, Nambara E, Nakajima M, Kawashima M, Satou M, Kim JM, Kobayashi N, Toyoda T, Shinozaki K, Seki M (2008) Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol 49(8):1135–1149. https://doi.org/10.1093/pcp/pcn101
Meng C, Cai C, Zhang T, Guo W (2009) Characterization of six novel NAC genes and their responses to abiotic stresses in Gossypium hirsutum L. Plant Sci 176(3):352–359. https://doi.org/10.1016/j.plantsci.2008.12.003
Mengjun Wu LG (2018) TCseq: time course sequencing data analysis. R package version 1.6.1
Missihoun TD, Hou Q, Mertens D, Bartels D (2014) Sequence and functional analyses of the aldehyde dehydrogenase 7B4 gene promoter in Arabidopsis thaliana and selected Brassicaceae: regulation patterns in response to wounding and osmotic stress. Planta 239(6):1281–1298. https://doi.org/10.1007/s00425-014-2051-0
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59(1):651–681. https://doi.org/10.1146/annurev.arplant.59.032607.092911
Myers C, Romanowsky SM, Barron YD, Garg S, Azuse CL, Curran A, Davis RM, Hatton J, Harmon AC, Harper JF (2009) Calcium-dependent protein kinases regulate polarized tip growth in pollen tubes. Plant J 59(4):528
Olías R, Eljakaoui Z, Li J, De Morales PA, Marín Manzano MC, Pardo JM, Belver A (2009) The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ 32(7):904–916. https://doi.org/10.1111/j.1365-3040.2009.01971.x
Pang C, Wang B (2008) Oxidative stress and salt tolerance in plants. Springer, Berlin, pp 231–245
Pardo JM (2010) Biotechnology of water and salinity stress tolerance. Curr Opin Biotechnol 21(2):185–196. https://doi.org/10.1016/j.copbio.2010.02.005
Peng Z, He S, Gong W, Sun J, Pan Z, Xu F, Lu Y, Du X (2014) Comprehensive analysis of differentially expressed genes and transcriptional regulation induced by salt stress in two contrasting cotton genotypes. BMC Genomics 15:760. https://doi.org/10.1186/1471-2164-15-760
Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL (2016) Transcript-level expression analysis of RNA-seq experiments with HISAT StringTie and Ballgown. Nat Protoc 11(9):1650–1667. https://doi.org/10.1038/nprot.2016.095
Qi Z, Spalding EP (2004) Protection of plasma membrane K+ transport by the salt overly sensitive1 Na+-H+ antiporter during salinity stress. Plant Physiol 136(1):2548–2555. https://doi.org/10.1104/pp.104.049213
Qin LX, Zhang DJ, Huang GQ, Li L, Li J, Gong SY, Li XB, Xu WL (2013) Cotton GhHyPRP3 encoding a hybrid proline-rich protein is stress inducible and its overexpression in Arabidopsis enhances germination under cold temperature and high salinity stress conditions. Acta Physiol Plant 35(5):1531–1542
Ramegowda V, Senthil-Kumar M (2015) The interactive effects of simultaneous biotic and abiotic stresses on plants: mechanistic understanding from drought and pathogen combination. J Plant Physiol 176:47–54. https://doi.org/10.1016/j.jplph.2014.11.008
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. https://doi.org/10.1093/bioinformatics/btp616
Rock CO, Calder RB, Karim MA, Jackowski S (2000) Pantothenate kinase regulation of the intracellular concentration of coenzyme A. J Biol Chem 275(2):1377–1383
Rodriguez-Uribe L, Higbie SM, Stewart JM, Wilkins T, Lindemann W, Sengupta-Gopalan C, Zhang J (2011) Identification of salt responsive genes using comparative microarray analysis in Upland cotton (Gossypium hirsutum L.). Plant Sci 180(3):461–469
Rubio S, Whitehead L, Larson TR, Graham IA, Rodriguez PL (2008) The coenzyme A biosynthetic enzyme phosphopantetheine adenylyltransferase plays a crucial role in plant growth, salt/osmotic stress resistance, and seed lipid storage. Plant Physiol 148(1):546–556. https://doi.org/10.1104/pp.108.124057
Schulz P, Herde M, Romeis T (2013) Calcium-dependent protein kinases: hubs in plant stress signaling and development. Plant Physiol 163(2):523–530. https://doi.org/10.1104/pp.113.222539
Schuster J, Binder S (2005) The mitochondrial branched-chain aminotransferase (AtBCAT-1) is capable to initiate degradation of leucine, isoleucine and valine in almost all tissues in Arabidopsis thaliana. Plant Mol Biol 57(2):241–254. https://doi.org/10.1007/s11103-004-7533-1
Shan C, Mei Z, Duan J, Chen H, Feng H, Cai W (2014) OsGA2ox5, a gibberellin metabolism enzyme, is involved in plant growth, the root gravity response and salt stress. PLoS One 9(1):e87110. https://doi.org/10.1371/journal.pone.0087110
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. https://doi.org/10.1101/gr.1239303
Shavrukov Y (2012) Salt stress or salt shock: which genes are we studying? J Exp Bot 64(1):119–127. https://doi.org/10.1093/jxb/ers316
Shi W, Hao L, Li J, Liu D, Guo X, Li H (2014) The Gossypium hirsutum WRKY gene GhWRKY39-1 promotes pathogen infection defense responses and mediates salt stress tolerance in transgenic Nicotiana benthamiana. Plant Cell Rep 33(3):483–498. https://doi.org/10.1007/s00299-013-1548-5
Slonim DK, Yanai I (2009) Getting started in gene expression microarray analysis. PLoS Comput Biol 5(10):e1000543
Soderman E, Mattsson J, Engstrom P (1996) The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. Plant J 10(2):375–381. https://doi.org/10.1046/j.1365-313X.1996.10020375.x
Sui N, Tian S, Wang W, Wang M, Fan H (2017) Overexpression of glycerol-3-phosphate acyltransferase from Suaeda salsa improves salt tolerance in Arabidopsis. Front Plant Sci. https://doi.org/10.3389/fpls.2017.01337
Sunarpi HT, Motoda J, Kubo M, Yang H, Yoda K, Horie R, Chan W, Leung H, Hattori K, Konomi M, Osumi M, Yamagami M, Schroeder JI, Uozumi N (2005) Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J 44(6):928–938. https://doi.org/10.1111/j.1365-313X.2005.02595.x
Vaidyanathan H, Sivakumar P, Chakrabarty R, Thomas G (2003) Scavenging of reactive oxygen species in NaCl-stressed rice (Oryza sativa L.)—differential response in salt-tolerant and sensitive varieties. Plant Sci 165(6):1411–1418. https://doi.org/10.1016/j.plantsci.2003.08.005
Wang G, Zhu Q, Meng Q, Wu C (2012) Transcript profiling during salt stress of young cotton (Gossypium hirsutum) seedlings via Solexa sequencing. Acta Physiol Plant 34(1):107–115. https://doi.org/10.1007/s11738-011-0809-6
Wei Y, Xu Y, Lu P, Wang X, Li Z, Cai X, Zhou Z, Wang Y, Zhang Z, Lin Z, Liu F, Wang K (2017) Salt stress responsiveness of a wild cotton species (Gossypium klotzschianum) based on transcriptomic analysis. PLoS One 12(5):e178313. https://doi.org/10.1371/journal.pone.0178313
Werckreichhart D, Feyereisen R (2000) Cytochromes P450: a success story. Genome Biol 1(6):1–9
Wiese J, Kranz T, Schubert S (2004) Induction of pathogen resistance in barley by abiotic stress. Plant Biol 6(5):529–536. https://doi.org/10.1055/s-2004-821176
Wu C, Yang G, Meng Q, Zheng C (2004) The cotton GhNHX1 gene encoding a novel putative tonoplast Na+/H+ antiporter plays an important role in salt stress. Plant Cell Physiol 45(5):600–607. https://doi.org/10.1093/pcp/pch071
Xu J, Tian Y, Peng R, Xiong A, Zhu B, Jin X, Gao F, Fu X, Hou X, Yao Q (2010) AtCPK6, a functionally redundant and positive regulator involved in salt/drought stress tolerance in Arabidopsis. Planta 232(4):1007. https://doi.org/10.1007/s00425-010-1216-8
Xue T, Li X, Zhu W, Wu C, Yang G, Zheng C (2009) Cotton metallothionein GhMT3a, a reactive oxygen species scavenger, increased tolerance against abiotic stress in transgenic tobacco and yeast. J Exp Bot 60(1):339
Yan H, Jia H, Chen X, Hao L, An H, Guo X (2014) The cotton WRKY transcription factor GHWRKY17 functions in drought and salt stress in transgenic Nicotianabenthamiana through ABA signalling and the modulation of reactive oxygen species production. Plant Cell Physiol 55(12):2060–2076
Zhang D, Yang H, Jia W, Huang C (2001) Protein phosphorylation is involved in the water stress-induced ABA accumulation in the roots ofMalus hupehensis Rehd. Chin Sci Bull 46(10):855–858. https://doi.org/10.1007/BF02900438
Zhang X, Zhen J, Li Z, Kang D, Yang Y, Kong J, Hua J (2011) Expression profile of early responsive genes under salt stress in upland cotton (Gossypium hirsutum L.). Plant Mol Biol Rep 29(3):626–637
Zhang YM, Zhang HM, Liu ZH, Li HC, Guo XL, Li GL (2015) The wheat NHX antiporter gene TaNHX2 confers salt tolerance in transgenic alfalfa by increasing the retention capacity of intracellular potassium. Plant Mol Biol 87(3):317–327. https://doi.org/10.1007/s11103-014-0278-6
Zhang F, Zhu G, Du L, Shang X, Cheng C, Yang B, Hu Y, Cai C, Guo W (2016) Genetic regulation of salt stress tolerance revealed by RNA-Seq in cotton diploid wild species Gossypium davidsonii. Sci Rep 6(1):20582. https://doi.org/10.1038/srep20582
Zhao J, Li S, Jiang T, Liu Z, Zhang W, Jian G, Qi F (2012) Chilling stress–the key predisposing factor for causing Alternaria alternata infection and leading to cotton (Gossypium hirsutum L.) leaf senescence. PLoS One 7(4):e36126. https://doi.org/10.1371/journal.pone.0036126
Zhao J, Gao Y, Zhang Z, Chen T, Guo W, Zhang T (2013) A receptor-like kinase gene (GbRLK) from Gossypium barbadense enhances salinity and drought-stress tolerance in Arabidopsis. BMC Plant Biol 13(1):110
Acknowledgements
This work was sponsored by Financial Foundation of Hebei Academy of Agriculture and Forestry Sciences (F17C2017039345), Innovation project of Hebei Academy of Agriculture and Forestry Sciences (2019-1-3-2), and Hebei Science and Technology Plan Project (16226303D). We would like to thank Lesley Benyon, PhD, from Liwen Bianji, Edanz Group China (https://www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Y. Wang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11738_2020_3117_MOESM1_ESM.tif
Supplementary file1 Supplementary Figure S1 The effect of exposure to 200 mM NaCl on the H15 and ZM12 varieties of upland cotton. (TIF 5543 kb)
11738_2020_3117_MOESM2_ESM.tif
Supplementary file2 Supplementary Figure S2 Comparison of the expression of 12 randomly selected genes by using RNA-seq and qRT-PCR. The gene expression values by qRT-PCR were transformed to log2 scale. (TIF 4407 kb)
11738_2020_3117_MOESM3_ESM.tif
Supplementary file3 Supplementary Figure S3 Venn diagram analysis of the numbers of DE genes in two cotton genotypes under different time points. (TIF 5271 kb)
11738_2020_3117_MOESM4_ESM.tif
Supplementary file4 Supplementary Figure S4 Heat map analysis of the expression levels of ROS genes based on log2 fold change. (TIF 7006 kb)
11738_2020_3117_MOESM7_ESM.xls
Supplementary file7 Supplementary Table S3 Differentially expressed genes after salt stress in the roots of H15. (XLS 7662 kb)
11738_2020_3117_MOESM8_ESM.xls
Supplementary file8 Supplementary Table S4 Differentially expressed genes after salt stress in the roots of ZM12. (XLS 2819 kb)
11738_2020_3117_MOESM9_ESM.xls
Supplementary file9 Supplementary Table S5 Detailed information of quantitative RT-PCR results and DESeq2 results. (XLS 26 kb)
11738_2020_3117_MOESM12_ESM.xls
Supplementary file12 Supplementary Table S8. H15-specific differentially expressed genes encoding CyP450 enzymes. (XLS 42 kb)
11738_2020_3117_MOESM14_ESM.xls
Supplementary file14 Supplementary Table S10. H15-specific differentially expressed genes for coenzyme A metabolic process. (XLS 24 kb)
Rights and permissions
About this article
Cite this article
Wang, Y., Liu, J., Zhao, G. et al. Dynamic transcriptome and co-expression network analysis of the cotton (Gossypium hirsutum) root response to salinity stress at the seedling stage. Acta Physiol Plant 42, 143 (2020). https://doi.org/10.1007/s11738-020-03117-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-020-03117-w