Abstract
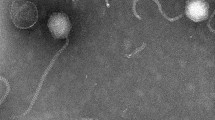
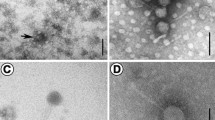
Although Alteromonas is ubiquitous in the marine environment, very little is known about Alteromonas phages, with only ten, thus far, being isolated and reported on. In this study, a novel double-stranded DNA phage, Alteromonas phage P24, which infects Alteromonas macleodii, was isolated from the coastal waters off Qingdao. Alteromonas phage P24 has a siphoviral morphology, with an icosahedral head, 61 ± 1 nm in diameter, and a tail length of 105 ± 1 nm. Alteromonas phage P24 contains lipids. It has an optimal temperature and pH for growth of 20℃ and 5–7, respectively. A one-step growth curve shows a latent period of 55 min, a rise period of 65 min, and an average burst size of approximately 147 virions per cell. Alteromonas phage P24 has the genome of 46,945 bp with 43.80% GC content and 74 open reading frames (ORFs) without tRNA. The results of the phylogenetic tree, based on the mcp and terL genes, show that Alteromonas phage P24 is closely related to Aeromonas phage phiARM81ld. Meanwhile, phylogenetic analysis based on the whole genome of P24 indicates that it forms a unique viral sub-cluster within Siphoviridae. This study contributes to the understanding of the genomic characteristics and the virus-host interactions of Alteromonas phages.




Similar content being viewed by others
Data Availability
The complete genome sequence of Alteromonas phage P24 was submitted to the GenBank under accession number MK241539. BioSample accession numbers (SAMN11618747) and BioProject accession numbers (PRJNA527681) are available in the NCBI BioSample database (https://www.ncbi.nlm.nih.gov/biosample/) and BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/), respectively.
References
Suttle CA (2005) 03—Reading—Viruses in the sea. Nature 437:356–361. https://doi.org/10.1038/nature04160
Gobler CJ, Hutchins DA, Fisher NS et al (1997) Release and bioavailability of C, N, P, Se, and Fe following viral lysis of a marine chrysophyte. Limnol Oceanogr 42:1492–1504. https://doi.org/10.4319/lo.1997.42.7.1492
Fuhrman J, Suttle C (1993) Viruses in marine planktonic systems. Oceanography 6:51–63. https://doi.org/10.5670/oceanog.1993.14
Fuhrman JA (1999) Marine viruses and their biogeochemical and ecological effects. Nature 399:541–548. https://doi.org/10.1038/21119
Suttle CA (1994) The significance of viruses to mortality in aquatic microbial communities. Microb Ecol 28:234–237. https://doi.org/10.1007/BF00166813
Baumann L, Baumann P, Mandel M, Allen RD (1972) Taxonomy of aerobic marine eubacteria. J Bacteriol. https://doi.org/10.1128/jb.110.1.402-429.1972
López-Pérez M, Gonzaga A, Martin-Cuadrado AB et al (2012) Genomes of surface isolates of Alteromonas macleodii: The life of a widespread marine opportunistic copiotroph. Sci Rep 2:1–11. https://doi.org/10.1038/srep00696
Romera-Castillo C, Sarmento H, Alvarez-Salgado XAÁ et al (2011) Net production and consumption of fluorescent colored dissolved organic matter by natural bacterial assemblages growing on marine phytoplankton exudates. Appl Environ Microbiol 77:7490–7498. https://doi.org/10.1128/AEM.00200-11
García-Martínez J, Acinas SG, Massana R, Rodríguez-Valera F (2002) Prevalence and microdiversity of Alteromonas macleodii-like microorganisms in different oceanic regions. Environ Microbiol 4:42–50. https://doi.org/10.1046/j.1462-2920.2002.00255.x
Pukall R, Buntefuß D, Frühling A et al (1999) Sulfitobacter mediterraneus sp. nov., a new sulfite-oxidizing member of the α-Proteobacteria. Int J Syst Bacteriol 49:513–519. https://doi.org/10.1099/00207713-49-2-513
Gao Y, Liu Q, Wang M et al (2017) Characterization and genome sequence of marine Alteromonas gracilis phage PB15 isolated from the Yellow Sea, China. Curr Microbiol 74:821–826. https://doi.org/10.1007/s00284-017-1251-9
Garcia-Heredia I, Rodriguez-Valera F, Martin-Cuadrado A-B (2013) Novel group of podovirus infecting the marine bacterium Alteromonas macleodii. Bacteriophage 3:e24766. https://doi.org/10.4161/bact.24766
Haq IU, Chaudhry WN, Andleeb S, Qadri I (2012) Isolation and partial characterization of a virulent bacteriophage IHQ1 specific for Aeromonas punctata from stream water. Microb Ecol 63:954–963. https://doi.org/10.1007/s00248-011-9944-2
Li Y, Wang M, Liu Q et al (2016) Complete genomic sequence of bacteriophage H188: a novel Vibrio kanaloae phage isolated from Yellow Sea. Curr Microbiol 72:628–633. https://doi.org/10.1007/s00284-015-0984-6
Kwiatek M, Parasion S, Rutyna P et al (2017) Isolation of bacteriophages and their application to control Pseudomonas aeruginosa in planktonic and biofilm models. Res Microbiol 168:194–207. https://doi.org/10.1016/j.resmic.2016.10.009
Kauffman KM, Hussain FA, Yang J et al (2018) A major lineage of non-tailed dsDNA viruses as unrecognized killers of marine bacteria. Nature 554:118–122. https://doi.org/10.1038/nature25474
Wang D, Jiang Y, Xiao S et al (2019) Characterization and genome analysis of a novel Alteromonas phage JH01 isolated from the Qingdao Coast of China. Curr Microbiol. https://doi.org/10.1007/s00284-019-01751-3
Capra ML, Quiberoni A, Reinheimer JA (2004) Thermal and chemical resistance of Lactobacillus casei and Lactobacillus paracasei bacteriophages. Lett Appl Microbiol 38:499–504. https://doi.org/10.1111/j.1472-765X.2004.01525.x
Pajunen M, Kiljunen S, Skurnik M (2000) Bacteriophage φYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J Bacteriol 182:5114–5120. https://doi.org/10.1128/JB.182.18.5114-5120.2000
Liu Y, Zhao L, Wang M et al (2019) Complete genomic sequence of bacteriophage P23: a novel Vibrio phage isolated from the Yellow Sea. China Virus Genes. https://doi.org/10.1007/s11262-019-01699-3
Gong Z, Wang M, Yang Q et al (2017) Isolation and complete genome sequence of a novel Pseudoalteromonas phage PH357 from the Yangtze River Estuary. Curr Microbiol 74:832–839. https://doi.org/10.1007/s00284-017-1244-8
Liu Z, Wang M, Meng X et al (2017) Isolation and genome sequencing of a novel Pseudoalteromonas phage PH1. Curr Microbiol 74:1–7. https://doi.org/10.1007/s00284-016-1175-9
Wojciak JM, Sarkar D, Landy A, Clubb RT (2002) Arm-site binding by λ-integrase: solution structure and functional characterization of its amino-terminal domain. Proc Natl Acad Sci USA 99:3434–3439. https://doi.org/10.1073/pnas.052017999
Groth AC, Olivares EC, Thyagarajan B, Calos MP (2000) A phage integrase directs efficient site-specific integration in human cells. Proc Natl Acad Sci USA 97:5995–6000. https://doi.org/10.1073/pnas.090527097
Fogg PCM, Haley JA, Marshall Stark W, Smith MCM (2017) Genome integration and excision by a new Streptomyces bacteriophage, φJoe. Appl Environ Microbiol. https://doi.org/10.1128/AEM.02767-16
Pell LG, Kanelis V, Donaldson LW et al (2009) The phage λ major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc Natl Acad Sci USA 106:4160–4165. https://doi.org/10.1073/pnas.0900044106
Medina E, Wieczorek D, Medina EM et al (2010) Assembly and maturation of the bacteriophage lambda procapsid: GpC Is the viral protease. J Mol Biol 401:813–830. https://doi.org/10.1016/j.jmb.2010.06.060
Sullivan MB, Coleman ML, Quinlivan V et al (2008) Portal protein diversity and phage ecology. Environ Microbiol 10:2810–2823. https://doi.org/10.1111/j.1462-2920.2008.01702.x
Newcomb WW, Thomsen DR, Homa FL, Brown JC (2003) Assembly of the herpes simplex virus capsid: identification of soluble scaffold-portal complexes and their role in formation of portal-containing capsids. J Virol 77:9862–9871. https://doi.org/10.1128/jvi.77.18.9862-9871.2003
Andrews BT, Catalano CE (2012) The enzymology of a viral genome packaging motor is influenced by the assembly state of the motor subunits. Biochemistry. https://doi.org/10.1021/bi300890y
Catalano CE, Cue D, Feiss M (1995) Virus DNA packaging: the strategy used by phage λ. Mol Microbiol 16:1075–1086. https://doi.org/10.1111/j.1365-2958.1995.tb02333.x33
Trudil D (2015) Phage lytic enzymes: a history. Virol Sin 30:26–32. https://doi.org/10.1007/s12250-014-3549-0
Wion D, Casadesús J (2006) N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat Rev Microbiol 4:183–192. https://doi.org/10.1038/nrmicro1350
García-Del Portillo F, Pucciarelli MG, Casadesús J (1999) DNA adenine methylase mutants of Salmonella typhimurium show defects in protein secretion, cell invasion, and M cell cytotoxicity. Proc Natl Acad Sci USA 96:11578–11583. https://doi.org/10.1073/pnas.96.20.11578
Nathans D, Smith HO (1975) Restriction endonucleases in the analysis and restructuring of DNA molecules. Annu Rev Biochem 44:273–293. https://doi.org/10.1146/annurev.bi.44.070175.001421
Kalatzis PG, Rørbo N, Castillo D et al (2017) Stumbling across the same phage: comparative genomics of widespread temperate phages infecting the fish pathogen Vibrio anguillarum. Viruses 9:122. https://doi.org/10.3390/v9050122
Murphy J, Mahony J, Ainsworth S et al (2013) Bacteriophage orphan DNA methyltransferases: Insights from their bacterial origin, function, and occurrence. Appl Environ Microbiol 79:7547–7555. https://doi.org/10.1128/mmbr.00011-16
Casjens SR (2005) Comparative genomics and evolution of the tailed-bacteriophages. Curr Opin Microbiol 8:451–458. https://doi.org/10.1016/j.mib.2005.06.014
Pečenková T, Pačes V (1999) Molecular phylogeny of Φ29-like phages and their evolutionary relatedness to other protein-primed replicating phages and other phages hosted by gram-positive bacteria. J Mol Evol 48:197–208. https://doi.org/10.1007/PL00006458
Kallies R, Kiesel B, Zopfi J et al (2017) Complete Genome Sequence of Alteromonas Virus vB_AspP-H4/4. Genome Announcements. https://doi.org/10.1128/genomea.00914-17
Acknowledgments
This study was financially supported by the Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (No.2018SDKJ0406-6), National Key Research and Development Program of China: 2018YFC1406704, The Fundamental Research Funds for the Central Universities (201812002), Natural Science Foundation of China(No. 41606153; 41976117; 41676178; 41906126).
Author information
Authors and Affiliations
Contributions
Yantao Liang, Hongbing Shao, Yong Jiang, and Min Wang designed the experiments and critically evaluated the manuscript. Xinran Zhang analyzed the data and wrote the manuscript. Yundan Liu isolated the phage and conducted the biological characterization experiments. Meiwen Wang, Tong Jiang, Jianhua Sun, and Chen Gao conducted the sequencing experiments. Andrew McMinn and Cui Guo critically revised and polished the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, X., Liu, Y., Wang, M. et al. Characterization and Genome Analysis of a Novel Marine Alteromonas Phage P24. Curr Microbiol 77, 2813–2820 (2020). https://doi.org/10.1007/s00284-020-02077-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02077-1