Abstract
Planctomycetes are ubiquitous bacteria with environmental and biotechnological relevance. Axenic cultures of planctomycetal strains are the basis to analyse their unusual biology and largely uncharacterised metabolism in more detail. Here, we describe strain Mal4T isolated from marine sediments close to Palma de Mallorca, Spain. Strain Mal4T displays common planctomycetal features, such as division by polar budding and the presence of fimbriae and crateriform structures on the cell surface. Cell growth was observed at ranges of 10–39 °C (optimum at 31 °C) and pH 6.5–9.0 (optimum at 7.5). The novel strain shows as pear-shaped cells of 2.0 ± 0.2 × 1.4 ± 0.1 µm and is one of the rare examples of orange colony-forming Planctomycetes. Its genome has a size of 7.7 Mb with a G+C content of 63.4%. Phylogenetically, we conclude that strain Mal4T (= DSM 100296T = LMG 29133T) is the type strain representing the type species of a novel genus, for which we propose the name Maioricimonas rarisocia gen. nov., sp. nov.
Similar content being viewed by others
Introduction
Planctomycetes are bacteria that belong to the PVC superphylum (Wagner and Horn 2006), which includes the phyla Planctomycetes, Verrucomicrobia, Chlamydiae, Lentisphaerae and Kirimatiellaeota as well as some uncultured candidate phyla, such as Candidatus Omnitrophica. The PVC superphylum has environmental, medical and biotechnological relevance (Devos and Ward 2014).
Planctomycetes have been shown to be present in several environments, in which they play important roles in biogeochemical cycles, such as the carbon and nitrogen cycle (Wiegand et al. 2018). One example are Planctomycetes of the class Candidatus Brocadiae, which perform unique reactions during anaerobic ammonium oxidation (anammox) (Strous et al. 1999; Peeters and van Niftrik 2019). Members of the phylum Planctomycetes, in particular of the class Planctomycetia, colonise a variety of environments from terrestrial to aquatic, being able to dwell on various marine algal surfaces (Bengtsson et al. 2012; Bondoso et al. 2014, 2015, 2017; Lage and Bondoso 2014; Vollmers et al. 2017). They form biofilms on biotic surfaces (Bengtsson and Øvreås 2010), on which they metabolise complex carbon substrates (Lachnit et al. 2013; Jeske et al. 2013). Unique pili-forming crateriform structures and an enlarged periplasm are probably required for uptake and also cleavage of large polysaccharides obtained from the environment (Boedeker et al. 2017).
Planctomycetes possess large genomes with sizes of up to 12.4 Mb (Ravin et al. 2018), in which the presence of giant genes has been reported (Jeske et al. 2013; Guo et al. 2014; Kohn et al. 2016; Faria et al. 2018). These genome sizes are in line with their assumed capacity for secondary metabolite production (Graça et al. 2016; Jeske et al. 2016; Yadav et al. 2018). Furthermore, several members of the phylum Planctomycetes produce carotenoids, which could be associated with an increased tolerance against UV radiation or oxidative stress (Kallscheuer et al. 2019b).
Planctomycetes were considered exceptional due to several presumptively eukaryotic features, such as the lack of a peptidoglycan (König et al. 1984), a compartmentalised cell plan (Lindsay et al. 1997), a nucleus-like structure (Fuerst and Webb 1991) and the endocytosis-like uptake of macromolecules for an intracellular degradation (Lonhienne et al. 2010). However, with advances of microscopy techniques and the development of genetic tools (Jogler et al. 2011; Rivas-Marín et al. 2016b; Boedeker et al. 2017), many of these traits have been refuted or reinterpreted.
In recent years, the presence of peptidoglycan has been reported in several members of the Planctomycetes (Jeske et al. 2015; van Teeseling et al. 2015) and also in the sister phyla Verrucomicrobia (Rast et al. 2017) and Chlamydiae (Pilhofer et al. 2013; Liechti et al. 2014, 2016). With the exception of anammox-performing Planctomycetes (Jogler 2014; Neumann et al. 2014), the proposed cell plan has been found to feature large invaginations of the cytoplasmic membrane instead of closed compartments (Santarella-Mellwig et al. 2013; Acehan et al. 2014; Boedeker et al. 2017). These discoveries contributed to the reinterpretation of Planctomycetes as bacteria with a cell envelope architecture resembling that of Gram-negative bacteria, but with some variations (Devos 2014a, b; Boedeker et al. 2017). Nevertheless, Planctomycetes remain exceptional in other ways, e.g. they lack the protein FtsZ normally essential for bacterial division as well as other division proteins (Pilhofer et al. 2008; Jogler et al. 2012; Rivas-Marín et al. 2016a). Beyond that, phylum members divide by binary fission, budding or intermediate mechanisms (Wiegand et al. 2018, 2020). Presence and essentiality of sterols in the membranes of one of its members was recently reported (Pearson et al. 2003; Rivas-Marin et al. 2019).
The unusual cell biology of Planctomycetes prompted us to explore the uncharacterised planctomycetal diversity. In the present study, we describe the novel strain Mal4T isolated from marine sediments in Palma de Mallorca (Spain) in terms of physiological, microscopic as well as genomic properties. Supported by phylogenetic analyses, we conclude that strain Mal4T represents a novel species of a novel genus within the family Planctomycetaceae.
Materials and methods
Cultivation conditions and isolation
Strain Mal4T was isolated from marine sediments at the coast of S’Arenal close to Palma de Mallorca (Spain) on the 23th of September 2014 (sampling location: 39.5126 N 2.7470 E) as previously described (Wiegand et al. 2020). For strain isolation and cultivation M1H NAG ASW medium was used. Medium preparation was previously described (Kallscheuer et al. 2019a). Cultures were incubated in baffled flasks at 28 °C with constant agitation at 110 rpm. Plates were cultivated at 28 °C for 2–3 weeks and isolated colonies were then streaked on fresh M1H NAG ASW plates. Initial amplification and sequencing of the 16S rRNA gene, intended to check whether isolated strains are members of the phylum Planctomycetes, was performed as previously described (Rast et al. 2017).
Physiological analyses
Cultivations for physiological assays were performed in M1H NAG ASW medium. For pH optimum determination, 100 mM 2-(N-morpholino)ethanesulfonic acid (MES) was used for cultivations at pH 5.0, 5.5, 6.0 and 6.5. For cultivations at pH values ranging from 7.0 to 8.0, MES was replaced by 100 mM 4-(2-hydroxyethyl)-1-piperazine-ethanesulfonic acid (HEPES), whereas 100 mM 3-(4-(2-Hydroxyethyl)piperazin-1-yl)-propane-1-sulfonic acid (HEPPS) served as a buffering agent at pH 8.5 and 100 mM N-cyclohexyl-2-aminoethanesulfonic acid (CHES) was used for pH maintenance at pH 9.0 and 9.5. Cultivations for determination of the pH optimum were performed at 28 °C. For temperature optimum determination, strain Mal4T was cultivated at pH 8.0 at different temperatures ranging from 10 to 39 °C. Cell growth and maximal growth rates were inferred from optical density measurements at 600 nm (OD600) of triplicate cultures.
Genome analysis
The genome of strain Mal4T was previously published (Wiegand et al. 2020). The genome (accession number CP036275) and 16S rRNA gene sequence (accession number MK559979) are available from the GenBank database. The primary metabolism was analysed by examining locally computed InterProScan (Mitchell et al. 2019) results cross-referenced with information from the UniProt database (UniProt 2019) and BlastP results of ‘typical’ protein sequences. Numbers of carbohydrate-active enzymes were determined by employing dbCAN2 (Zhang et al. 2018), which automatically mines the CAZy database (Lombard et al. 2014).
Light microscopy and scanning electron microscopy
Phase contrast microscopy and scanning electron microscopy were performed according to protocols published earlier (Kallscheuer et al. 2019a).
Phylogenetic analyses
16S rRNA gene sequence-based phylogeny was computed for strain Mal4T, the type strains of all described planctomycetal species (assessed in January 2020) and all isolates recently published (Kohn et al. 2016, 2020a, b; Boersma et al. 2019; Kallscheuer et al. 2019a; Dedysh et al. 2020; Wiegand et al. 2020). An alignment of 16S rRNA gene sequences was performed with SINA (Pruesse et al. 2012). The phylogenetic analysis was conducted employing a maximum likelihood approach with 1000 bootstraps, the nucleotide substitution model GTR, gamma distribution and estimation of proportion of invariable sites (GTRGAMMAI option) (Stamatakis 2014). Three 16S rRNA genes of bacterial strains from the PVC superphylum, but outside of the phylum Planctomycetes, were used as outgroup. The rpoB nucleotide sequences (encoding the RNA polymerase β-subunit) were taken from publicly available genome annotations and the sequence identities were determined as described previously (Bondoso et al. 2013) using Clustal Omega (Sievers et al. 2011). Alignment and matrix calculation were done after extracting only those parts of the sequence that would have been sequenced with the described primer set. The average nucleotide identity (ANI) was calculated using OrthoANI (Lee et al. 2016). The average amino acid identity (AAI) was obtained with the aai.rb script of the enveomics collection (Rodriguez-R and Konstantinidis 2016). The percentage of conserved proteins (POCP) was calculated as described before (Qin et al. 2014). The unique single-copy core genome of all analysed genomes for the multi-locus sequence analysis (MLSA) was determined with proteinortho5 (Lechner et al. 2011) (‘selfblast’ option enabled). The sequences of the obtained orthologous groups were aligned using MUSCLE v.3.8.31 (Edgar 2004). After clipping, partially aligned C- and N-terminal regions and poorly aligned internal regions were filtered using Gblocks (Castresana 2000). The final alignment was concatenated and clustered using the maximum likelihood method implemented by RaxML (Stamatakis 2014) with the ‘rapid bootstrap’ method and 500 bootstrap replicates.
Results and discussion
Phylogenetic inference
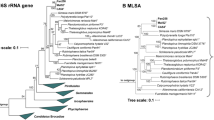
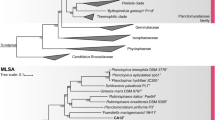
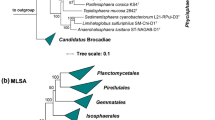
In the phylogenetic trees obtained after analysis of 16S rRNA gene sequences, as well as MLSA (Fig. 1), strain Mal4T clusters stably with members of two genera of the family Planctomycetaceae, namely Planctomicrobium and Thalassoglobus. 16S rRNA gene sequence identity between strain Mal4T and the two genera is between 91.4% and 91.9% (Fig. 2). These values are below the proposed genus threshold of 94.5%, but above the threshold for separate families of 86.5% (Yarza et al. 2014), indicating that strain Mal4T represents an distinct genus in the family Planctomycetaceae. Coherently, average nucleotide identities (ANI) below 95% confirm that strain Mal4T is a distinct species. Phylogenetic assumptions on the genus level can also be obtained by analysing the rpoB gene sequence identities, AAI and POCP. For delineation of genera, the proposed threshold values for the above-mentioned markers are 75.5–78% (Kallscheuer et al. 2019c), 45–65% (Konstantinidis et al. 2017) and 50% (Qin et al. 2014), respectively. The rpoB identity value and the AAI between strain Mal4T and the members of the genus Thalassoglobus, which comprises Thalassoglobus neptunius KOR42T (Kohn et al. 2020a) and Thalassoglobus polymorphus Mal48T (Rivas-Marin et al. 2020), are below the given thresholds. POCP was found to be slightly above the threshold (51.2%), this, however, does not significantly influence the overall conclusion that strain Mal4T belongs to a separate genus. Minimal comparative values of strain Mal4T and the genus Rubinisphaera, another closely related genus; featuring Rubinisphaera italica (Kallscheuer et al. 2019a) and Rubinisphaera brasiliensis (Scheuner et al. 2014), are below these thresholds for all three phylogenetic markers (Fig. 2). Analogously, POCP between strain Mal4T and Planctomicrobium piriforme P3T (Kulichevskaya et al. 2015) was also found to fall below the proposed threshold (Fig. 2), whilst the AAI value (56%) was in a ‘grey zone’ (45–65%), but well below the upper limit. Although the rpoB gene sequence identity of 78.6% is slightly above the proposed threshold, this sole deviance should not overrule the distinctiveness of the other values. In summary, the majority of analysed phylogenetic markers suggests that strain Mal4T belongs to a novel genus.
Similarity values of the novel isolate Mal4T in relation to species P. piriforme P3T, Thalassoglobus sp. and Rubinisphaera sp. Methods used: 16S rRNA gene sequence identity, rpoB gene identity, percentage of conserved proteins (POCP), average nucleotide identity (ANI) and average amino acid identity (AAI)
Morphological and physiological analyses
Light microscopy and scanning electron microscopy (Fig. 3) were applied to analyse the morphological characteristic of Mal4T cells harvested during the exponential growth phase. Detailed information on morphology and cell division is summarised in comparison to the current closest relatives (Table 1). Mal4T cells are pear-shaped (2.0 ± 0.2 µm × 1.4 ± 0.1 µm) (Fig. 3a–c), occur as single cells and in rare cases form aggregates (Fig. 3d). The cell surface appears rough, evenly covered with crateriform structures and short fimbriae (Fig. 3d, e). A holdfast structure was not observed during electron microscopy. As shown for all described members of the family Planctomycetaceae, cell division takes place by polar budding with the daughter cell displaying a round shape. Optimal temperature and pH for growth were shown to be 31 °C and pH 7.5, respectively, however, Mal4T cells are able to grow over a range of 10–39 °C and pH 6.5–9.0 (Fig. 4). These values are comparable to the two Thalassoglobus species, but differ from P. piriforme P3T, which did not grow at temperatures exceeding 30 °C and favours moderate acidic conditions. The maximal observed growth rate of strain Mal4T in M1H NAG ASW medium was determined to be 0.041 h−1, corresponding to a generation time of approximately 17 h. Strain Mal4T is amongst the rare examples of planctomycetal strains forming orange colonies and might thus be an interesting strain for further analysis of carotenoid production and their function in Planctomycetes. Since most of the planctomycetal strains characterised so far are either pink to red or lack pigmentation (white), the pigmentation of the novel strain is an important phenotypic feature separating it from its current close phylogenetic neighbours. Strain Mal4T is an aerobic heterotroph.
Microscopy images and cell size plot of strain Mal4T. Pictures from light microscopy (a, b) and scanning electron microscopy (d, e) are shown. The scale bars are 1 µm. For determination of the cell size (c) at least 100 representative cells were counted manually or by using a semi-automated object count tool
Genomic characteristics
The genomic characteristics of strain Mal4T in comparison to T. polymorphus Mal48T, T. neptunius KOR42T and P. piriforme P3T are summarised in Table 1. Its genome is 7.7 Mb in size, which is around 1 Mb larger compared to the other three strains. The G+C content is also the highest of the four strains. Automated gene prediction and annotation identified 5829 putative protein-encoding genes, of which 39% (2257 genes) are annotated as hypothetical proteins. These values correspond to 753 protein-coding genes per Mb and a coding density of 85.9%. Although the genome size of strain Mal4T is larger, the coding density is in the same range in the other three species. Similar to its relatives, the strain lacks plasmids. Numbers of 41–55 tRNA genes are similar, except for T. neptunius KOR42T, which has a slightly higher number of 70 tRNA genes. As for T. polymorphus Mal48T, strain Mal4T harbours two copies of the 16S rRNA gene, whereas the gene occurs in single copy in the other two strains.
Genome-encoded features of the primary carbon metabolism
Based on the genome sequences, we analysed key metabolic capabilities in the primary metabolism of strain Mal4T in comparison to the two Thalassoglobus species and P. piriforme P3T (Table 2). Genes coding for enzymes participating in glycolytic pathways, gluconeogenesis, the tricarboxylic acid (TCA) cycle and anaplerotic reactions, such as pyruvate or phosphoenolpyruvate carboxylation and the glyoxylate shunt, were included. The resulting data suggest that strain Mal4T is able to metabolise carbohydrates using at least two glycolytic pathways, the Embden-Meyerhof-Parnas pathway (the most common glycolytic pathway) and the pentose phosphate pathway. Additionally, its genome bears genes coding for putative 2-dehydro-3-deoxyphosphogluconate aldolase and phosphogluconate dehydratase, both involved in the alternative Entner-Doudoroff pathway. All four strains harbour a complete gene set required for a functional TCA cycle, which suggests that the central carbon metabolism of the strains is similar to canonical heterotrophic bacteria. With regard to gluconeogenesis, a minimal gene set required for this pathway has been identified, suggesting that the three strains are capable of de novo sugar biosynthesis. All four strains lack the glyoxylate shunt, which is typically required during growth either with acetate or with compounds that are degraded to acetate or acetyl-CoA units. The lack of the glyoxylate shunt suggests that the strains are not able to use such compounds as the exclusive energy and carbon source. Alternatively, they may follow a different pathway with a similar function.
Based on the physiological, morphological and phylogenetic analyses of strain Mal4T, we conclude that the characterised strain represents a novel species within the novel genus Maioricimonas. Thus, we propose the name Maioricimonas rarisocia gen. nov., sp. nov., represented by the type strain Mal4T.
Maioricimonas gen. nov.
Maioricimonas (Ma.io.ri.ci’mo.nas. M.L. fem. n. Maiorica of Mallorca; L. fem. n. monas a unit, monad; N.L. fem. n. Maioricimonas a monad from Mallorca, Spain).
Cells have a Gram-negative cell envelope architecture and divide by polar budding. Cells are mesophilic, neutrophilic, aerobic and heterotrophic and present crateriform structures and matrix or fimbriae. The genus is part of the family Planctomycetaceae, order Planctomycetales, class Planctomycetia, phylum Planctomycetes. The type species of the genus is Maioricimonas rarisocia.
Maioricimonas rarisocia sp. nov.
Maioricimonas rarisocia (ra.ri.so’ci.a. L. masc. adj. rarus few, infrequent; L. masc. adj. socius allied, united; N.L. fem. adj. rarisocia; corresponding to the characteristic of the cells to seldom form aggregates).
In addition to the features described for the genus, cells are pear-shaped (2.0 × 1.4 µm), form orange colonies and mostly occur as single cells. Temperature and pH optimum of the type strain are 31 °C and 7.5, respectively, however growth is observed over a range of 10–39 °C and pH 6.5–9.0. The type strain genome (accession number CP036275) and 16S rRNA gene (accession number MK559979) are available from GenBank. The genome of the type strain has a G+C content of 63.4% and a size of 7.7 Mb.
The type strain is Mal4T (= DSM 100296T = LMG 29133T, deposited as strain Malle4), isolated from marine sediments near the coast of S’Arenal in Palma de Mallorca, Mallorca Island, Spain.
References
Acehan D, Santarella-Mellwig R, Devos DP (2014) A bacterial tubulovesicular network. J Cell Sci 127:277. https://doi.org/10.1242/jcs.137596
Bengtsson MM, Øvreås L (2010) Planctomycetes dominate biofilms on surfaces of the kelp Laminaria hyperborea. BMC Microbiol 10:261. https://doi.org/10.1186/1471-2180-10-261
Bengtsson MM, Sjøtun K, Lanzén A, Ovreås L (2012) Bacterial diversity in relation to secondary production and succession on surfaces of the kelp Laminaria hyperborea. ISME J 6:2188–2198. https://doi.org/10.1038/ismej.2012.67
Boedeker C, Schüler M, Reintjes G et al (2017) Determining the bacterial cell biology of Planctomycetes. Nat Commun 8:14853. https://doi.org/10.1038/ncomms14853
Boersma AS, Kallscheuer N, Wiegand S et al (2019) Alienimonas californiensis gen. nov. sp. nov., a novel Planctomycete isolated from the kelp forest in Monterey Bay. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01367-4
Bondoso J, Albuquerque L, Nobre MF et al (2015) Roseimaritima ulvae gen. nov., sp. nov. and Rubripirellula obstinata gen. nov., sp. nov. two novel planctomycetes isolated from the epiphytic community of macroalgae. Syst Appl Microbiol 38:8–15. https://doi.org/10.1016/j.syapm.2014.10.004
Bondoso J, Balagué V, Gasol JM, Lage OM (2014) Community composition of the Planctomycetes associated with different macroalgae. FEMS Microbiol Ecol 88:445–456. https://doi.org/10.1111/1574-6941.12258
Bondoso J, Godoy-Vitorino F, Balagué V et al (2017) Epiphytic Planctomycetes communities associated with three main groups of macroalgae. FEMS Microbiol Ecol 93:fiw255. https://doi.org/10.1093/femsec/fiw255
Bondoso J, Harder J, Lage OM (2013) rpoB gene as a novel molecular marker to infer phylogeny in Planctomycetales. Antonie Van Leeuwenhoek 104:477–488. https://doi.org/10.1007/s10482-013-9980-7
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. https://doi.org/10.1093/oxfordjournals.molbev.a026334
Dedysh SN, Henke P, Ivanova AA et al (2020) 100-year-old enigma solved: identification, genomic characterization and biogeography of the yet uncultured Planctomyces bekefii. Environ Microbiol 22:198–211
Devos DP (2014a) PVC bacteria: variation of, but not exception to, the Gram-negative cell plan. Trends Microbiol 22:14–20. https://doi.org/10.1016/j.tim.2013.10.008
Devos DP (2014b) Re-interpretation of the evidence for the PVC cell plan supports a Gram-negative origin. Antonie Van Leeuwenhoek 105:271–274. https://doi.org/10.1007/s10482-013-0087-y
Devos DP, Ward NL (2014) Mind the PVCs. Environ Microbiol 16:1217–1221. https://doi.org/10.1111/1462-2920.12349
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Faria M, Bordin N, Kizina J et al (2018) Planctomycetes attached to algal surfaces: insight into their genomes. Genomics 110:231–238. https://doi.org/10.1016/j.ygeno.2017.10.007
Fuerst JA, Webb RI (1991) Membrane-bounded nucleoid in the eubacterium Gemmata obscuriglobus. Proc Natl Acad Sci USA 88:8184–8188. https://doi.org/10.1073/pnas.88.18.8184
Graça AP, Calisto R, Lage OM (2016) Planctomycetes as novel source of bioactive molecules. Front Microbiol 7:1241. https://doi.org/10.3389/fmicb.2016.01241
Guo M, Zhou Q, Zhou Y et al (2014) Genomic evolution of 11 type strains within family Planctomycetaceae. PLoS ONE 9:e86752. https://doi.org/10.1371/journal.pone.0086752
Jeske O, Jogler M, Petersen J et al (2013) From genome mining to phenotypic microarrays: Planctomycetes as source for novel bioactive molecules. Antonie Van Leeuwenhoek 104:551–567. https://doi.org/10.1007/s10482-013-0007-1
Jeske O, Schüler M, Schumann P et al (2015) Planctomycetes do possess a peptidoglycan cell wall. Nat Commun 6:7116
Jeske O, Surup F, Ketteniß M et al (2016) Developing techniques for the utilization of Planctomycetes as producers of bioactive molecules. Front Microbiol 7:1242. https://doi.org/10.3389/fmicb.2016.01242
Jogler C (2014) The bacterial “mitochondrium”. Mol Microbiol 94:751–755. https://doi.org/10.1111/mmi.12814
Jogler C, Glöckner FO, Kolter R (2011) Characterization of Planctomyces limnophilus and development of genetic tools for its manipulation establish it as a model species for the phylum Planctomycetes. Appl Environ Microbiol 77:5826–5829. https://doi.org/10.1128/AEM.05132-11
Jogler C, Waldmann J, Huang X et al (2012) Identification of proteins likely to be involved in morphogenesis, cell division, and signal transduction in Planctomycetes by comparative genomics. J Bacteriol 194:6419–6430. https://doi.org/10.1128/JB.01325-12
Kallscheuer N, Jogler M, Wiegand S et al (2019a) Rubinisphaera italica sp. nov. isolated from a hydrothermal area in the Tyrrhenian Sea close to the volcanic island Panarea. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01329-w
Kallscheuer N, Moreira C, Airs R et al (2019b) Pink- and orange-pigmented Planctomycetes produce saproxanthin-type carotenoids including a rare C45 carotenoid. Environ Microbiol Rep 11:741–748. https://doi.org/10.1111/1758-2229.12796
Kallscheuer N, Wiegand S, Peeters SH et al (2019c) Description of three bacterial strains belonging to the new genus Novipirellula gen. nov., reclassificiation of Rhodopirellula rosea and Rhodopirellula caenicola and readjustment of the genus threshold of the phylogenetic marker rpoB for Planctomycetaceae. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01374-5
Kohn T, Heuer A, Jogler M et al (2016) Fuerstia marisgermanicae gen. nov., sp. nov., an unusual member of the phylum Planctomycetes from the German Wadden Sea. Front Microbiol 7:2079. https://doi.org/10.3389/fmicb.2016.02079
Kohn T, Rast P, Wiegand S et al (2020a) The microbiome of Posidonia oceanica seagrass leaves can be dominated by Planctomycetes. Front Microbiol 11:1458. https://doi.org/10.3389/fmicb.2020.01458
Kohn T, Wiegand S, Boedeker C et al (2020b) Planctopirus ephydatiae, a novel Planctomycete isolated from a freshwater sponge. Syst Appl Mirobiol 43:126022. https://doi.org/10.1016/j.syapm.2019.126022
König E, Schlesner H, Hirsch P (1984) Cell wall studies on budding bacteria of the Planctomyces/Pasteuria group and on a Prosthecomicrobium sp. Arch Microbiol 138:200–205. https://doi.org/10.1007/BF00402120
Konstantinidis KT, Rosselló-Móra R, Amann R (2017) Uncultivated microbes in need of their own taxonomy. ISME J 11:2399–2406. https://doi.org/10.1038/ismej.2017.113
Kulichevskaya IS, Ivanova AA, Detkova EN et al (2015) Planctomicrobium piriforme gen. nov., sp. nov., a stalked planctomycete from a littoral wetland of a boreal lake. Int J Syst Evol Microbiol 65:1659–1665. https://doi.org/10.1099/ijs.0.000154
Lachnit T, Fischer M, Künzel S et al (2013) Compounds associated with algal surfaces mediate epiphytic colonization of the marine macroalga Fucus vesiculosus. FEMS Microbiol Ecol 84:411–420. https://doi.org/10.1111/1574-6941.12071
Lage OM, Bondoso J (2014) Planctomycetes and macroalgae, a striking association. Front Microbiol 5:267. https://doi.org/10.3389/fmicb.2014.00267
Lechner M, Findeiß S, Steiner L et al (2011) Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics 12:124. https://doi.org/10.1186/1471-2105-12-124
Lee I, Ouk Kim Y, Park S-C, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103. https://doi.org/10.1099/ijsem.0.000760
Liechti G, Kuru E, Packiam M et al (2016) Pathogenic Chlamydia lack a classical sacculus but synthesize a narrow, mid-cell peptidoglycan ring, regulated by MreB, for cell division. PLoS Pathog 12:e1005590–e1005590. https://doi.org/10.1371/journal.ppat.1005590
Liechti GW, Kuru E, Hall E et al (2014) A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506:507–510. https://doi.org/10.1038/nature12892
Lindsay M, Webb R, Fuerst J (1997) Pirellulosomes: a new type of membrane-bounded cell compartment in Planctomycete bacteria of the genus Pirellula. Microbiology 143:739–748
Lombard V, Golaconda Ramulu H, Drula E et al (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. https://doi.org/10.1093/nar/gkt1178
Lonhienne TGA, Sagulenko E, Webb RI et al (2010) Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci USA 107:12883–12888. https://doi.org/10.1073/pnas.1001085107
Mitchell AL, Attwood TK, Babbitt PC et al (2019) InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res 47:D351–D360. https://doi.org/10.1093/nar/gky1100
Neumann S, Wessels HJCT, Rijpstra WIC et al (2014) Isolation and characterization of a prokaryotic cell organelle from the anammox bacterium Kuenenia stuttgartiensis. Mol Microbiol 94:794–802. https://doi.org/10.1111/mmi.12816
Pearson A, Budin M, Brocks JJ (2003) Phylogenetic and biochemical evidence for sterol synthesis in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci USA 100:15352–15357. https://doi.org/10.1073/pnas.2536559100
Peeters SH, van Niftrik L (2019) Trending topics and open questions in anaerobic ammonium oxidation. Curr Opin Chem Biol 49:45–52. https://doi.org/10.1016/j.cbpa.2018.09.022
Pilhofer M, Aistleitner K, Biboy J et al (2013) Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat Commun 4:2856
Pilhofer M, Rappl K, Eckl C et al (2008) Characterization and evolution of cell division and cell wall synthesis genes in the bacterial phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and phylogenetic comparison with rRNA genes. J Bacteriol 190:3192–3202. https://doi.org/10.1128/JB.01797-07
Pruesse E, Peplies J, Glöckner FO (2012) SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829. https://doi.org/10.1093/bioinformatics/bts252
Qin Q-L, Xie B-B, Zhang X-Y et al (2014) A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol 196:2210–2215. https://doi.org/10.1128/JB.01688-14
Rast P, Glöckner I, Boedeker C et al (2017) Three novel species with peptidoglycan cell walls form the new genus Lacunisphaera gen. nov. in the family Opitutaceae of the verrucomicrobial subdivision 4. Front Microbiol 8:202. https://doi.org/10.3389/fmicb.2017.00202
Ravin NV, Rakitin AL, Ivanova AA et al (2018) Genome analysis of fimbriiglobus ruber SP5T, a planctomycete with confirmed chitinolytic capability. Appl Environ Microbiol 84:e02645-17
Rivas-Marín E, Canosa I, Devos DP (2016a) Evolutionary cell biology of division mode in the bacterial Planctomycetes-Verrucomicrobia-Chlamydiae superphylum. Front Microbiol 7:1964. https://doi.org/10.3389/fmicb.2016.01964
Rivas-Marín E, Canosa I, Santero E, Devos DP (2016b) Development of genetic tools for the manipulation of the planctomycetes. Front Microbiol. https://doi.org/10.3389/fmicb.2016.00914
Rivas-Marin E, Stettner S, Gottshall EY et al (2019) Essentiality of sterol synthesis genes in the planctomycete bacterium Gemmata obscuriglobus. Nat Commun 10:2916. https://doi.org/10.1038/s41467-019-10983-7
Rivas-Marin E, Wiegand S, Kallscheuer N et al (2020) Thalassoglobus polymorphus sp. nov., a novel Planctomycete isolated close to a public beach of Mallorca Island. Antonie van Leeuwenhoek. https://doi.org/10.1007/s10482-020-01437-y
Rodriguez-R LM, Konstantinidis KT (2016) The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints 4:e1900v1. https://doi.org/10.7287/peerj.preprints.1900v1
Santarella-Mellwig R, Pruggnaller S, Roos N et al (2013) Three-dimensional reconstruction of bacteria with a complex endomembrane system. PLoS Biol 11:e1001565. https://doi.org/10.1371/journal.pbio.1001565
Scheuner C, Tindall BJ, Lu M et al (2014) Complete genome sequence of Planctomyces brasiliensis type strain (DSM 5305T), phylogenomic analysis and reclassification of Planctomycetes including the descriptions of Gimesia gen. nov., Planctopirus gen. nov. and Rubinisphaera gen. nov. and emended descriptions of the order Planctomycetales and the family Planctomycetaceae. Stand Genomic Sci 9:10. https://doi.org/10.1186/1944-3277-9-10
Sievers F, Wilm A, Dineen D et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. https://doi.org/10.1038/msb.2011.75
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Strous M, Kuenen JG, Jetten MSM (1999) Key physiology of anaerobic ammonium oxidation. Appl Environ Microbiol 65:3248–3250
van Teeseling MCF, Mesman RJ, Kuru E et al (2015) Anammox Planctomycetes have a peptidoglycan cell wall. Nat Commun 6:6878. https://doi.org/10.1038/ncomms7878
Vollmers J, Frentrup M, Rast P et al (2017) Untangling genomes of novel Planctomycetal and verrucomicrobial species from monterey bay kelp forest metagenomes by refined binning. Front Microbiol 8:472. https://doi.org/10.3389/fmicb.2017.00472
Wagner M, Horn M (2006) The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol 17:241–249. https://doi.org/10.1016/j.copbio.2006.05.005
Wiegand S, Jogler M, Boedeker C et al (2020) Cultivation and functional characterization of 79 planctomycetes uncovers their unique biology. Nat Microbiol 5:126–140. https://doi.org/10.1038/s41564-019-0588-1
Wiegand S, Jogler M, Jogler C (2018) On the maverick Planctomycetes. FEMS Microbiol Rev 42:739–760. https://doi.org/10.1093/femsre/fuy029
Yadav S, Vaddavalli R, Siripuram S et al (2018) Planctopirus hydrillae sp. nov., an antibiotic producing Planctomycete isolated from the aquatic plant Hydrilla and its whole genome shotgun sequence analysis. J Antibiot 71:575–583. https://doi.org/10.1038/s41429-018-0035-1
Yarza P, Yilmaz P, Pruesse E et al (2014) Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12:635–645. https://doi.org/10.1038/nrmicro3330
Zhang H, Yohe T, Huang L et al (2018) dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46:W95–W101. https://doi.org/10.1093/nar/gky418
Acknowledgements
Open Access funding provided by Projekt DEAL. Part of this research was funded by the Deutsche Forschungsgemeinschaft (DFG) Grants KA 4967/1-1 and JO 893/4-1, Grant ALWOP.308 of the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO), SIAM (Soehngen Institute for Anaerobic Microbiology) Grant No. 024002002 and the Radboud Excellence fellowship. We thank Ina Schleicher for skillful technical assistance. We thank Brian Tindall and Regine Fähnrich from the DSMZ, as well as staff of the BCCM/LMG Bacteria collection, for support during strain deposition.
Author information
Authors and Affiliations
Contributions
E.R.M. wrote the manuscript, analysed the data and prepared the figures, S.W. and M.J. performed the genomic and phylogenetic analysis, A.H. isolated the strain and performed the initial strain cultivation and deposition, S.H.P. and C.B. performed the light microscopic analysis, N.K. and M.S.M.J. contributed to text preparation and revised the manuscript, M.R. performed the electron microscopic analysis, C.J. took the samples, supervised A.H. and the study. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rivas-Marin, E., Wiegand, S., Kallscheuer, N. et al. Maioricimonas rarisocia gen. nov., sp. nov., a novel planctomycete isolated from marine sediments close to Mallorca Island. Antonie van Leeuwenhoek 113, 1901–1913 (2020). https://doi.org/10.1007/s10482-020-01436-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-020-01436-z