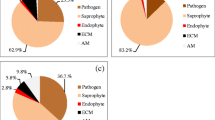
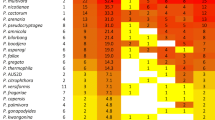
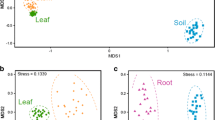
Abstract
Roots act as a biological filter that exclusively allows only a portion of the soil-associated microbial diversity to infect the plant. This microbial diversity includes organisms both beneficial and detrimental to plants. Phytophthora species are among the most important groups of detrimental microbes that cause various soil-borne plant diseases. We used a metabarcoding approach with Phytophthora-specific primers to compare the diversity and richness of Phytophthora species associated with roots of native and non-native trees, using different types of soil inocula collected from native and managed forests. Specifically, we analysed (1) roots of two non-native tree species (Eucalyptus grandis and Acacia mearnsii) and native trees, (2) roots of two non-native tree species from an in vivo plant baiting trial, (3) roots collected from the field versus those from the baiting trial, and (4) roots and soil samples collected from the field. The origin of the soil and the interaction between root and soil significantly influenced Phytophthora species richness. Moreover, species richness and community composition were significantly different between the field root samples and field soil samples with a higher number of Phytophthora species in the soil than in the roots. The results also revealed a substantial and previously undetected diversity of Phytophthora species from South Africa.




Similar content being viewed by others
References
Berendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17(8):478–486
Turner TR, James EK, Poole PS (2013) The plant microbiome. Genome Biol 14(6):1–10
Griffin EA, Carson WP (2018) Tree endophytes: cryptic drivers of tropical forest diversity. In: Pirttilä AM, Frank AC (eds) Endophytes of forest trees: biology and applications. Springer International Publishing, Cham, pp 63–103
Baldrian P (2017) Forest microbiome: diversity, complexity and dynamics. FEMS Microbiol Rev 41(2):109–130
Dickie IA, Bolstridge N, Cooper JA, Peltzer DA (2010) Co-invasion by Pinus and its mycorrhizal fungi. New Phytol 187(2):475–484
Ribeiro OK (2013) A historical perspective of Phytophthora. In: Lamour K (ed) Phytophthora: a global perspective, vol 2. CABI International, United Kingdom, pp. 1–10
Brasier CM, Cooke DE, Duncan JM, Hansen EM (2003) Multiple new phenotypic taxa from trees and riparian ecosystems in Phytophthora gonapodyides–P. megasperma ITS clade 6, which tend to be high-temperature tolerant and either inbreeding or sterile. Mycol Res 107(3):277–290
Marano AV, Jesus AL, De Souza JI, Jerônimo GH, Gonçalves DR, Boro MC, Rocha SCO, Pires-Zottarelli CLA (2016) Ecological roles of saprotrophic Peronosporales (Oomycetes, Straminipila) in natural environments. Fungal Ecol 19:77–88
Stamler RA, Sanogo S, Goldberg NP, Randall JJ (2016) Phytophthora species in rivers and streams of the southwestern United States. Appl Environ Microbiol 82(15):4696–4704
Crone M, McComb JA, O’Brien PA, Hardy GESJ (2013) Survival of Phytophthora cinnamomi as oospores, stromata, and thick-walled chlamydospores in roots of symptomatic and asymptomatic annual and herbaceous perennial plant species. Fungal Biol 117(2):112–123
Migliorini D, Ghelardini L, Tondini E, Luchi N, Santini A (2015) The potential of symptomless potted plants for carrying invasive soilborne plant pathogens. Divers Distrib 21(10):1218–1229
Pirozynski KA, Malloch DW (1975) The origin of land plants: a matter of mycotrophism. Biosystems 6(3):153–164
Selosse MA, Strullu-Derrien C, Martin FM, Kamoun S, Kenrick P (2015) Plants, fungi and oomycetes: a 400-million year affair that shapes the biosphere. New Phytol 206(2):501–506
Wager VA (1941) Descriptions of the south African Pythiaceae with records of their occurrence. Bothalia 4:3–35
Nagel JH, Gryzenhout M, Slippers B, Wingfield MJ (2013) The occurrence and impact of Phytophthora on the African continent. In: Lamour K (ed) Phytophthora: a global perspective, vol 2. CABI International, United Kingdom, pp. 204–214
Wingfield M, Knox-Davies P (1980) Observations on diseases in pine and eucalyptus plantations in South Africa. Phytophylactica 12(2):57–63
Roux J, Hurley B, Wingfield MJ, Bredenkamp BV, Upfold SJ (2012) Diseases and pests of eucalypts, pine and wattle. South African Forestry Handbook:303–335
Linde C, Kemp GHJ, Wingfield MJ (1994) Pythium and Phytophthora species associated with eucalypts and pines in South Africa. Eur J For Pathol 24(6–7):345–356
Bezuidenhout C, Denman S, Kirk S, Botha W, Mostert L, McLeod A (2010) Phytophthora taxa associated with cultivated Agathosma, with emphasis on the P. citricola complex and P. capensis sp. nov. Persoonia 25(1):32–49
Von Broembsen S (1984) Occurrence of Phytophthora cinnamomi on indigenous and exotic hosts in South Africa, with special reference to the South-Western Cape Province. Phytophylactica 16:221–225
van Wyk P (1973) Root and crown rot of silver trees. J S Afr Bot 39:255–260
Bose T, Wingfield MJ, Roux J, Vivas M, Burgess TI (2018) Community composition and distribution of Phytophthora species across adjacent native and non-native forests of South Africa. Fungal Ecol 36:17–25
Scott P, Burgess TI, Hardy GESJ (2013) Globalization and Phytophthora. In: Lamour K (ed) Phytophthora: a global perspective, vol 2. CABI International, United Kingdom, pp 226–232
Hulbert JM, Paap T, Burgess TI, Roets F, Wingfield MJ (2019) Botanical gardens provide valuable baseline Phytophthora diversity data. Urban For Urban Green 46:126461
Buee M, Reich M, Murat C, Morin E, Nilsson RH, Uroz S, Martin F (2009) 454 pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol 184(2):449–456
Tedersoo L, Bahram M, Toots M, Diedhiou AG, Henkel TW, Kjøller R, Morris MH, Nara K, Nouhra E, Peay KG (2012) Towards global patterns in the diversity and community structure of ectomycorrhizal fungi. Mol Ecol 21(17):4160–4170
Blaalid R, Carlsen T, Kumar S, Halvorsen R, Ugland KI, Fontana G, Kauserud H (2012) Changes in the root-associated fungal communities along a primary succession gradient analysed by 454 pyrosequencing. Mol Ecol 21(8):1897–1908
Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R, Ruiz LV, Vasco-Palacios AM, Thu PQ, Suija A (2014) Global diversity and geography of soil fungi. Science 346(6213):1256688
Burgess TI, McDougall KL, Scott PM, Hardy GESJ, Garnas J (2019) Predictors of Phytophthora diversity and community composition in natural areas across diverse Australian ecoregions. Ecography 42(3):565–577
Khaliq I, Hardy GESJ, White D, Burgess TI (2018) eDNA from roots: a robust tool for determining Phytophthora communities in natural ecosystems. FEMS Microbiol Ecol 94(5):fiy048
Burgess TI, White D, McDougall KM, Garnas J, Dunstan WA, Català S, Carnegie AJ, Worboys S, Cahill D, Vettraino A-M, Stukely MJC, Liew ECY, Paap T, Bose T, Migliorini D, Williams N, Brigg F, Crane C, Rudman T, Hardy GESJ (2017) Phytophthora distribution and diversity across Australia. Pac Conserv Biol 23(1):1–13
Català S, Berbegal M, Pérez-Sierra A, Abad-Campos P (2017) Metabarcoding and development of new real-time specific assays reveal Phytophthora species diversity in holm oak forests in eastern Spain. Plant Pathol 66(1):115–123. https://doi.org/10.1111/ppa.12541
Català S, Pérez-Sierra A, Abad-Campos P (2015) The use of genus-specific amplicon pyrosequencing to assess Phytophthora species diversity using eDNA from soil and water in northern Spain. PLoS One 10(3):e0119311
Prigigallo MI, Abdelfattah A, Cacciola SO, Faedda R, Sanzani SM, Cooke DE, Schena L (2016) Metabarcoding analysis of Phytophthora diversity using genus-specific primers and 454 pyrosequencing. Phytopathology 106(3):305–313
Riddell CE, Frederickson-Matika D, Armstrong AC, Elliot M, Forster J, Hedley PE, Morris J, Thorpe P, Cooke DE, Pritchard L (2019) Metabarcoding reveals a high diversity of woody host-associated Phytophthora spp. in soils at public gardens and amenity woodlands in Britain. PeerJ 7:e6931
Scibetta S, Schena L, Chimento A, Cacciola SO, Cooke DEL (2012) A molecular method to assess Phytophthora diversity in environmental samples. J Microbiol Methods 88(3):356–368
Katoh K, Misawa K, Ki K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30(14):3059–3066
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52(5):696–704
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9(8):772–772
R Core Team (2018) R: a language and environment for statistical computing. R Foundation for Statistical Computing. https://www.r-project.org/
Anderson MJ (2006) Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62(1):245–253
Anderson MJ, Walsh DCI (2013) PERMANOVA, ANOSIM, and the mantel test in the face of heterogeneous dispersions: what null hypothesis are you testing? Ecol Monogr 83(4):557–574
Abad ZG, Burgess TI, Bienapfl JC, Redford AJ, Coffey M, Knight L (2019) IDphy: molecular and morphological identification of Phytophthora based on the types. USDA APHIS PPQ S&T Beltsville lab, USDA APHIS PPQ S&T ITP, and Centre for Phytophthora Science and Management. http://idtools.org/id/phytophthora/index.php. Accessed 15 February 2020
Kamoun S (2001) Nonhost resistance to Phytophthora: novel prospects for a classical problem. Curr Opin Plant Biol 4(4):295–300
Nürnberger T, Lipka V (2005) Non-host resistance in plants: new insights into an old phenomenon. Mol Plant Pathol 6(3):335–345
Erwin DC, Ribeiro OK (1996) Phytophthora diseases worldwide. American Phytopathological Society, St. Paul, Minnesota
Jung T, Orlikowski L, Henricot B, Abad-Campos P, Aday A, Aguín Casal O, Bakonyi J, Cacciola S, Cech T, Chavarriaga D (2016) Widespread Phytophthora infestations in European nurseries put forest, semi-natural and horticultural ecosystems at high risk of Phytophthora diseases. For Pathol 46(2):134–163
Hardy GESJ, Sivasithamparam K (1988) Phytophthora spp. associated with container-grown plants in nurseries in Western Australia. Plant Dis 72(5):435–437
Parke JL, Grünwald NJ (2012) A systems approach for management of pests and pathogens of nursery crops. Plant Dis 96(9):1236–1244
Barber PA, Paap T, Burgess TI, Dunstan W, Hardy GESJ (2013) A diverse range of Phytophthora species are associated with dying urban trees. Urban For Urban Green 12(4):569–575
Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P (2013) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838
Soltis DE, Soltis PS, Chase MW, Mort ME, Albach DC, Zanis M, Savolainen V, Hahn WH, Hoot SB, Fay MF (2000) Angiosperm phylogeny inferred from 18S rDNA, rbcL, and atpB sequences. Bot J Linn Soc 133(4):381–461
Whitfield F, Shea S, Gillen K, Shaw K (1981) Volatile components from the roots of Acacia pulchella R. Br. and their effect on Phytophthora cinnamomi Rands. Aust J Bot 29(2):195–208
D'Souza NK, Colquhoun IJ, Shearer BL, Hardy GESJ (2004) The potential of five Western Australian native Acacia species for biological control of Phytophthora cinnamomi. Aust J Bot 52(2):267–279
Smith I, Marks G (1983) Influence of Acacia spp. on the control of Phytophthora cinnamomi root rot of Eucalyptus sieberi. Aust For Res 13(3–4):231–240
D’Souza N, Colquhoun I, Sheared B, Hardy GSJ (2005) Assessing the potential for biological control of Phytophthora cinnamomi by fifteen native Western Australian jarrah-forest legume species. Australas Plant Pathol 34(4):533–540
Kozanitas M, Osmundson TW, Linzer R, Garbelotto M (2017) Interspecific interactions between the sudden oak death pathogen Phytophthora ramorum and two sympatric Phytophthora species in varying ecological conditions. Fungal Ecol 28:86–96
Mazzola M (1998) Elucidation of the microbial complex having a causal role in the development of apple replant disease in Washington. Phytopathology 88(9):930–938
Cook RJ (2017) Untold stories: forty years of field research on root diseases of wheat. The American Phytopathological Society, St. Paul
Badri DV, Weir TL, van der Lelie D, Vivanco JM (2009) Rhizosphere chemical dialogues: plant–microbe interactions. Curr Opin Biotechnol 20(6):642–650
Babikova Z, Gilbert L, Bruce TJ, Birkett M, Caulfield JC, Woodcock C, Pickett JA, Johnson D (2013) Underground signals carried through common mycelial networks warn neighbouring plants of aphid attack. Ecol Lett 16(7):835–843
Pozo MJ, Azcón-Aguilar C (2007) Unraveling mycorrhiza-induced resistance. Curr Opin Plant Biol 10(4):393–398
Jung SC, Martinez-Medina A, Lopez-Raez JA, Pozo MJ (2012) Mycorrhiza-induced resistance and priming of plant defenses. J Chem Ecol 38(6):651–664
Blouin M (2018) Chemical communication: an evidence for co-evolution between plants and soil organisms. Appl Soil Ecol 123:409–415
Oh E, Gryzenhout M, Wingfield BD, Wingfield MJ, Burgess TI (2013) Surveys of soil and water reveal a goldmine of Phytophthora diversity in south African natural ecosystems. IMA Fungus 4(1):123–131
Nagel JH, Slippers B, Wingfield MJ, Gryzenhout M (2015) Multiple Phytophthora species associated with a single riparian ecosystem in South Africa. Mycologia 107(5):915–925
Hüberli D, Burgess TI, Hardy GESJ (2010) Fishing for Phytophthora across Western Australia’s waterways. In: 5th IUFRO Phytophthoras in forests and natural ecosystems, Auckland and Rotorua, New Zealand
Acknowledgements
The University of Pretoria, the Tree Protection Co-operative Programme (TPCP) and the Department of Science and Technology–National Research Foundation (DST-NRF) Centre of Excellence in Tree Health Biotechnology (CTHB) provided financial support for the present study. M.V. was supported by ‘Juan de la Cierva Program’, Ministry of Economy, Industry and Competitiveness, Government of Spain. We thank Mrs. Diane White of Murdoch University and Mrs. Frances Brigg from Western Australian State Agricultural Biotechnology Centre for their assistance with 454 sequencings. Sappi Ltd. and the Institute for Commercial Forestry Research (ICFR), Pietermaritzburg, South Africa provided seeds of E. grandis and A. mearnsii, respectively. We thank Mr. Jan Nagel, Mr. Conrad Trollip, Mr. Joey Hulbert and Ms. Mandy Messal, as well as foresters from NCT Forestry Co-operative Limited and TWK Agri for assistance with the fieldwork. Dr. Almuth Hammerbacher (FABI) provided substantial assistance in revising the manuscript, for which we are most grateful.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
FIG. S1. Soil and root collection sites across Mpumalanga and KwaZulu-Natal provinces of South Africa. (A, B) Soil and root samples were collected from four sites spanning across KwaZulu-Natal and Mpumalanga Provinces of South Africa. Close-up maps of the sampling sites at (C) Howick, (D) Vryheid, (E) Melmoth and (F) Commondale. For each close-up map of the collection sites, pointers in blue = Eucalyptus grandis, red = Acacia mearnsii and purple = natural forest. Bar equals to 100 m. FIG. S2. Flowchart for soil and root sampling at each collection sites. At each site, a total of 12 soil and root samples (six each) were collected. For the field-collected root samples, amplicon library was directly prepared from the total genomic DNA. Each soil sample was divided into two parts and baited with sterile grown seedlings of Eucalyptus grandis and Acacia mearnsii. After 5 months of incubation, the roots were harvested from each seedling followed by preparation of amplicon library. In total, we sequenced 72 samples (18 samples per collection site × 4 sites) from four collection sites. FIG. S3. Maximum-likelihood phylogenies using complete ITS1 gene region for Phytophthora species recovered from metabarcoding of root samples. The consensus sequence of MOTUs for each taxon was used for these five analyses. Suffix HTRSA indicates MOTUs recovered from the present study. Taxa names in pink font indicate new reports from South Africa. Numerical on the branches show bootstrap value > 70%. (DOCX 42451 kb)
ESM 2
Table S1. A list of Phytophthora species detected through environmental sequencing of soil and root samples from South Africa (present and a previous study (Bose et al. 2018)). Data for all first reports for formally described taxa were sourced from IDphy (https://idtools.org/id/phytophthora/). (DOCX 37 kb)
Rights and permissions
About this article
Cite this article
Bose, T., Wingfield, M.J., Roux, J. et al. Phytophthora Species Associated with Roots of Native and Non-native Trees in Natural and Managed Forests. Microb Ecol 81, 122–133 (2021). https://doi.org/10.1007/s00248-020-01563-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-020-01563-0