- Department of Life Sciences, Ben-Gurion University, Beer-Sheva, Israel
The discovery of Mariam, a wheat-unique miniature transposable element family, was reported in our previous study. We have also shown the possible impact of Mariam insertions on the expression of wheat genes. However, the evolutionary dynamics of Mariam was not studied in detail. In this study, we have assessed the insertion sites of Mariam family in different wheat species. In-silico analysis of Mariam insertions has allowed the discovery of two different sequence versions of Mariam, and that Mariam might have been recently active in wild emmer wheat genome (T. turgidum ssp diccocoides). In addition, the analysis of Mariam insertional polymorphism has facilitated the discovery of large genomic rearrangement events, such as deletions and introgressions in the wheat genome. The dynamics of Mariam family sheds light on the evolution of wheat.
Introduction
Transposable elements (TEs) are mobile DNA sequences that go through transposition, i.e., change their location within the genome (Kidwell, 2002). They are divided into two classes based on their mode of transposition: Class I—RNA elements, or Retrotransposons, that transpose through a “copy and paste” mechanism via RNA intermediate, and Class II—DNA elements that transpose through a “cut and paste” mechanism. In most class II elements, a transposase enzyme cuts the DNA transposon at the terminal inverted repeats (TIRs), excises the TE and inserts it into the target site (Sabot et al., 2004). Transposable elements are further divided into subclasses, orders, super-families and families based on the presence of different repeats, the proteins they use for transposition and the similarity of sequences. For example, in order to be considered from the same family, TEs have to share at least 80% sequence identity in at least 80% of their coding/internal sequence or their terminal repeats (Kidwell, 2002; Wicker et al., 2007).
TEs can be found in both prokaryotes and eukaryotes, and they have been found in all eukaryote species investigated so far. Some grasses such as maize, barley and wheat possess very high amount of TEs (Kidwell, 2002; Middleton et al., 2013). While the gene content in the wheat genome is only 2%, repetitive elements represent ~85% of the genome (Charles et al., 2008; Avni et al., 2017; Clavijo et al., 2017; Appels et al., 2018). Although TEs are usually silenced due to different host epigenetic regulations as DNA methylation, RNA interference and chromatin modification, they might be activated on behalf of different stresses (Slotkin and Martienssen, 2007; Dubin et al., 2018). Abiotic stresses, biotic stresses and genomic stresses such as hybridization or polyploidization can trigger activity of TEs (Capy et al., 2000; Kashkush et al., 2003; Levy and Feldman, 2004; Lisch, 2013; Keidar-Friedman et al., 2018). Transposition can have different genetic and epigenetics effects on the host genome (Casacuberta and Santiago, 2003; Mansour, 2007; Slotkin and Martienssen, 2007; Zhao et al., 2016).
Insertions into coding regions can cause mutations that might be harmful in some cases, and in others can give rise to new protein functions and even to domestication of TEs (Rebollo et al., 2012; Zhao et al., 2016; Schrader and Schmitz, 2018; Jiang et al., 2019). Insertions into non-coding sequences might affect the expression of a gene (Kashkush et al., 2003; Li et al., 2014; Dubin et al., 2018), alternative splicing and can even lead to exonization (Lev-Maor et al., 2003; Krull et al., 2005; Schmitz and Brosius, 2011; Dubin et al., 2018; Keidar et al., 2018). Transposable elements can also be involved or induce different chromosomal rearrangements such as inversions, duplications, deletions and translocations (Gray et al., 2000; Kidwell, 2002; Bariah et al., 2020).
Wheat is one of the most important crops in the world as it provides ~20% of the total calories consumed by humans. It also has a major role in the development of agriculture as it was one of first crops that were domesticated (Appels et al., 2018; Venske et al., 2019). The origin of wheat dates back to ~5 million years ago, when three diploid ancestral species were diverged from a common progenitor. These species include the donor of A genome—T. urartu, the donor of B genome—a close relative of today’s Ae. speltoides and the donor of D genome—Ae. tauschii.
~ 0.5 million years ago, a hybridization event between the donors of A and B sub-genomes was followed by polyploidization and generated the tetraploid wild emmer wheat (T. turgidum ssp diccocoides, AB). ~9,000 years ago, another hybridization event between the domesticated emmer wheat (AB, Triticum diccocon) and the donor of D genome, was followed by polyploidization and led to the speciation of the hexaploid bread wheat (ABD) (Levy and Feldman, 2002; Feldman and Levy, 2005; Petersen et al., 2006). Wild emmer wheat grows in populations across the Middle East Fertile Crescent, in different environmental conditions. It is considered based on molecular evidence that the tetraploidization event (speciation of wild emmer) occurred in the area of Mt. Hermon and the upper Jordan valley (Israel). In Israel, there are about 20 isolated or semi-isolated populations of wild emmer that grow between Mt. Hermon in the north to Mt. Amasa in the south (Nevo and Beiles, 1989; Volis et al., 2014).
In a previous study, we have reported on the discovery of Mariam, a wheat-unique miniature TE-like family that was found in the coding region of a gene that encodes for 5-formyltetrahydrofolate, in two accessions of wild emmer from Mt. Hermon population (Domb et al., 2019). In this study, we have studied the dynamics of Mariam family in wheat species, including insertional polymorphism in various wheat species, and their association with large genomic rearrangement events. We also observed a second version of Mariam family, which we termed Mariam2. The possible role of Mariam during wheat evolution is discussed.
Methods
Genomic Data
In this study, the most updated genome drafts of five Triticum and Aegilops species were used: (1) Triticum urartu (TU), accession G1812 (PI428198), the progenitor of wheat A sub-genome (https://www.ncbi.nlm.nih.gov/assembly/GCA_003073215.1) (Ling et al., 2018). (2) Aegilops tauschii ssp. strangulate (AT), accession AL8/78, the progenitor of wheat D sub-genome https://www.ncbi.nlm.nih.gov/assembly/GCA_000347335.2 (Luo et al., 2017). (3) Triticum turgidum ssp. diccocoides (WE), wild emmer wheat accession Zavitan, genome AB (assembly version 2. WEWseq: https://wheat.pw.usda.gov/graingenes_downloads/Zavitan/) (Avni et al., 2017). (4) Triticum aestivum (TA), bread wheat cultivar Chinese Spring, genome ABD, IWGSC RefSeq v2.0: https://urgi.versailles.inra.fr/download/iwgsc/IWGSC_RefSeq_Assemblies/ (Appels et al., 2018). (5) Triticum turgidum ssp. durum (DW), durum wheat cultivar Svevo, genome AB, https://www.ncbi.nlm.nih.gov/bioproject/518793 (Maccaferri et al., 2019).
Computer-Assisted Analysis of Mariam Insertions
Consensus sequences of the two versions; Mariam1 and Mariam2, were built with Multiple Sequence Alignment (MSA) using ClustalW algorithm in Ugene software (Okonechnikov et al., 2012) according to the full-length insertions. Mariam1 consensus sequence was built based on the 33 full-length insertions found in our previous study (Domb et al., 2019) and Mariam2 consensus sequence was built based on the full-length insertions found using MAK - MITE analysis kit (excluding A5-2, D6-2, and D7-1 insertions that were discovered later by the following analysis). MAK (http://labs.csb.utoronto.ca/yang/MAK/) is a homology-based software, it uses a consensus sequence as query and the BLASTN algorithm with global alignment. The consensus sequences (Supp. file 1 and Supp. file 2) were used each as a query sequence in NCBI Blast+ standalone version 2.6.0 (Camacho et al., 2009). BLASTN algorithm was used with an e-value of “1e-150,” best_hit_overhang command with value “0.25” in order to avoid replications of the same sequence and specific locations of hits were extracted (commands “sstart,” “sseq”). ClustalW algorithm was used for MSA, and phylogenetic trees were constructed using Maximum likelihood with Tamura-Nei model and 100 Bootsrap replications using MegaX software (Kumar et al., 2018). Venn diagram for common insertions between species was generated using the Bioinformatic & Evolutionary Genomics tools of Ghent University (http://bioinformatics.psb.ugent.be/webtools/Venn/), based on comparison of flanking sequences of Mariam insertions between the different species, as explained in the following section. All insertions, their specific location and TSD can be found in Table S1.
In Silico Analysis of Conserved Mariam Insertions, Their Associations With Genes and Genomic Rearrangements
Python scripts were used to retrieve Mariam insertions together with their flanking sequences according to their chromosomal locations, in order to examine target site duplications (TSD, in the range of 10 bp of Mariam flanking sequences), association with genes (in the range of 500 bp of Mariam flanking sequences), conservation among different wheat species (in the range of 1000 bp of Mariam flanking sequences) or involvement in genomic rearrangements (by analysis of ~5000 bp or more of Mariam flanking sequences). TSD was generated using WebLogo software (http://weblogo.berkeley.edu/logo.cgi). Each logo is composed of a stack of letters (nucleotides) for each nucleotide position in a sequence. Each letters’ height in the stack represents its relative frequency at the specific position, whereas stack width represents the relative fraction of nucleotides at that position.
In cases where the orthologous region (~1,000 bp of flanking sequences from both side of Mariam insertion) of an insertion site was polymorphic (presence vs. absence of a locus) among wheat species, chromosome walking analysis was performed to assesses possible genomic rearrangements events. Mariam-containing locus that was present in some species and absent in others was used as query in NCBI Blast+ BLASTN algorithm for chromosomal comparative analysis. If the BLASTN hits were too short or made of repetitive sequences, a larger region was used as query (by adding 10,000 bp or more to each side). In cases where no hits were found in the investigated chromosome, the whole genome of the investigated species was used as database. Following identification of the orthologues region, both regions were aligned to generate a dot plot in Ugene software v1.29.0 (Okonechnikov et al., 2012) using a minimum repeat length of 1000 bp and 90% repeat identity.
Plant Material and DNA Isolation
Accessions of wild diploid species—T. urartu (TU), Ae. searsii, Ae. speltoides and Ae. tauschii (AT), as well as accessions of the polyploid species—T. turgidum ssp durum (DW) and bread wheat (T. aestivum, TA) were used in this study. Seeds were kindly provided by Prof. Moshe Feldman, The Weizmann institute of Science, Rehovot, Israel. A collection of wild emmer wheat (T. turgidum ssp. dicoccoides, WE) populations from five geographically isolated sites in Israel was used in this study; Mt. Hermon, Amiad, Tabgha, Jaba and Mt. Amasa. The same collection was used in previous publications (Domb et al., 2017; Domb et al., 2019). See Table S2 for details about the plant accessions. Leaf material was harvested ~ 4 weeks post-germination. DNA was isolated using the DNeasy plant mini kit (Qiagen).
Site-Specific PCR
Primers for a subset of insert sites were designed in Primer3 software (See primer sequences in Table S3, http://bioinfo.ut.ee/primer3-0.4.0/) for PCR validation. Each PCR reaction was prepared with 10 μl of PCRBIO HS Taq Mix Red (PCRBiosystems), 7 μl of ultrapure water (Biological Industries), 1 μl of the site-specific primer (10 μM) and 1 μl of template genomic DNA (~ 50 ng/μl). The PCR conditions were 95°C incubation for 2 min, 40 cycles of 95°C for 10 s, the specific annealing temperature (calculated according to each primer set) for 15 s and 72°C for 15 s. PCR products were tested on 1.5% agarose gels and visualized with ethidium bromide (Amresco). Expected product sizes were determined by a DNA size standard (100 bp ladder, SMOBIO), and for some insertions the amplified products were extracted from gel and sequenced for validation.
Results
Analysis of Mariam Insertion Sites in Wheat Genome Drafts
Mariam is a family of miniature transposable elements (~300 bp in length), discovered in wild emmer wheat (T. turgidum ssp. dicoccoides), lacking TIRs (terminal inverted repeats) and other characteristics of known transposable elements (Domb et al., 2019). No hits were found for any known TE from both TREP (Wicker et al., 2002) and Repbase (Bao et al., 2015). Here, we have used the consensus sequence of Mariam elements found in our previous study to retrieve insertions from the publicly available wheat genome drafts. Full-length (~300 bp), as well as short versions (~93 - 240 bp) of Mariam insertions were detected. A total of 60 insertions were found in the 5 drafts; 3 insertions were found in T. urartu (TU), 4 in Ae. tauschii (AT), 12 in durum (DW), 21 in wild emmer (WE) and 20 insertions in bread wheat (TA).
Sequence comparison between full-length Mariam insertions within A, B and D sub-genomes of wheat, and between sub-genomes might provide insights into the mobility nature of Mariam elements, to examine whether Mariam transposition is made in a genome-specific manner. To this end, a Multiple Sequence Alignment (MSA) was performed on 21 Mariam insertions retrieved from wild emmer genome draft. A phylogenetic tree based on the MSA clustered insertions into 3 main groups (Figure 1A), while one insertion (B5-3S) was not clustered into any group. The 3 cluster groups consist of insertions from both A and B sub-genomes. These clusters might indicate possible transpositions between different chromosomes (such as the case of B4-6 and B5-6 insertions) and between different sub-genomes (such as cases; B1-4 and A7-3, A4-5 and B7-6 insertions). Similar analysis was performed on the 20 retrieved Mariam insertions from the bread wheat genome draft (Figure 1B). Here, the phylogenetic tree clustered insertions into 2 main groups, while one insertion (B3-1) was not clustered into any group. Within the 2 main groups, Mariam insertions were clustered based on: (1) chromosome-specific manner (e.g. insertions B1-1 and B1-2, insertions A6-5 and A6-1); (2) sub-genome-specific manner (e.g. insertions D3-1 and D4-1); or (3) none specific to sub-genome or chromosome (e.g. insertions D3-3 and A5-12S, insertions A4-2 and B6-4).
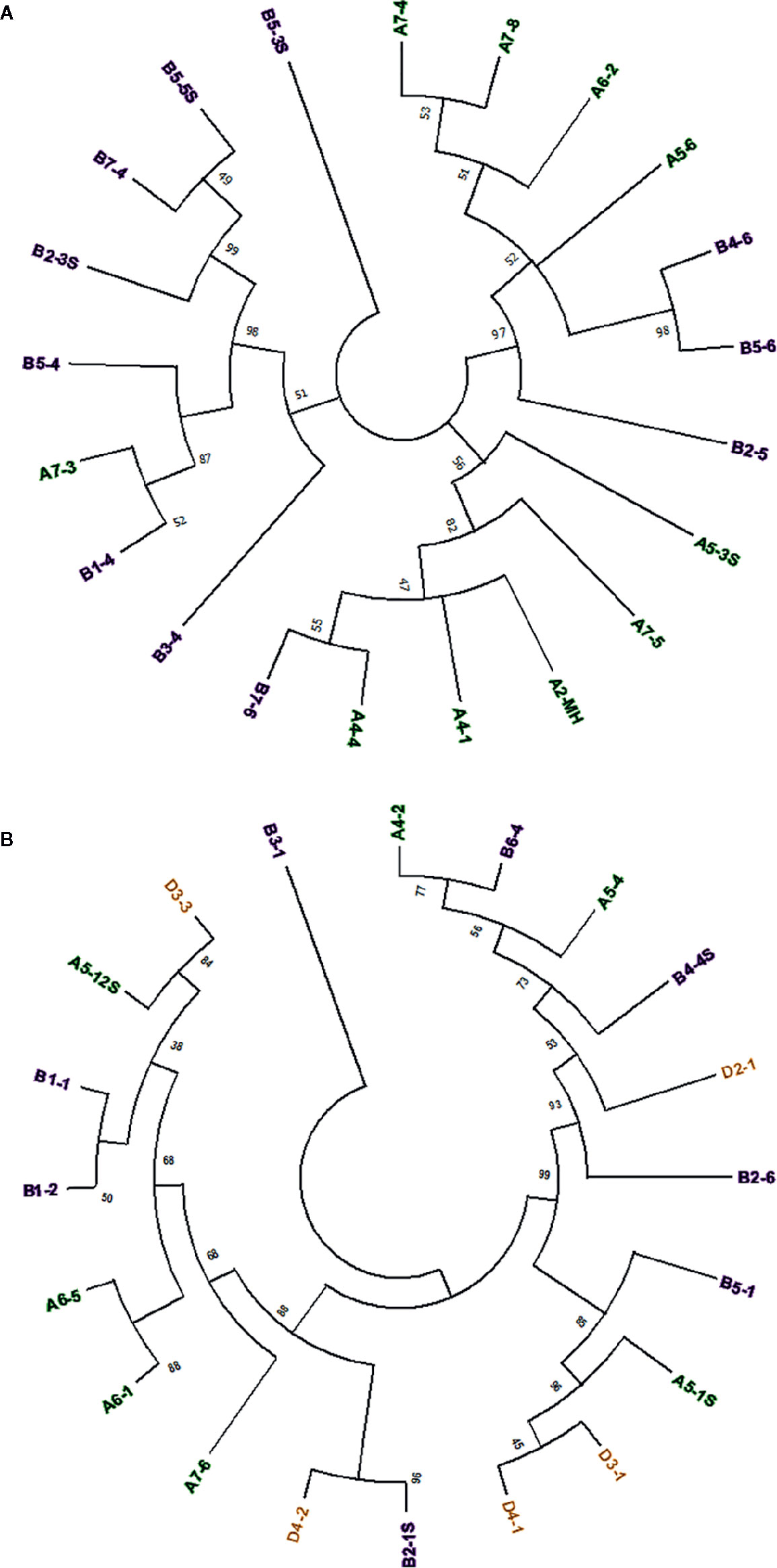
Figure 1 (A) Phylogenetic analysis of 21 Mariam sequences in wild emmer genome by Maximum Likelihood method. The insertions were clustered into three major groups and one insertion that was not clustered into any group (B5-3S) according to sequence similarity. (B) Phylogenetic analysis of 20 Mariam sequences in bread wheat genome by Maximum Likelihood method. The insertions were clustered into two major groups and one insertion that was not clustered into any group (B3-1) according to sequence similarity. Insertion codes are composed of the sub-genome (A/B/D), chromosome number (1-7) and a serial number. Insertions from A sub-genome are marked with green, insertions of B sub-genome are marked with purple and insertions of D sub-genome are marked with orange. A 45% cutoff parameter was used in both trees. The percentage of replicate trees in which the insertions clustered together in the bootstrap test (100 replicates) are shown next to the branches.
In order to assess the timing of Mariam proliferation during wheat evolution we have performed a comparative analysis of common Mariam insertions in the five genome drafts of Aegilops and Triticum species. Venn diagram (Figure 2) shows that: (1) only two Mariam insertions were common to all 5 species, indicating that those are ancient Mariam insertions; (2) six insertions were common to both Ae. tauschii (D) and bread wheat (ABD), indicating those were inherited from the D donor to bread wheat ~10,000 years ago; (3) ten insertions were common to allopolyploid species- durum, wild emmer and bread wheat and absent in diploid species, indicating that those might be accompanied with allopolyploidization process; (4) six insertions were unique to durum wheat, indicating specific Mariam proliferation during the lifetime of durum; (5) six insertions were unique to bread wheat, indicating specific Mariam proliferation during the lifetime of bread wheat; and (6) 15 insertions were unique to wild emmer wheat indicating relatively higher proliferation rates of Mariam in wild emmer wheat.
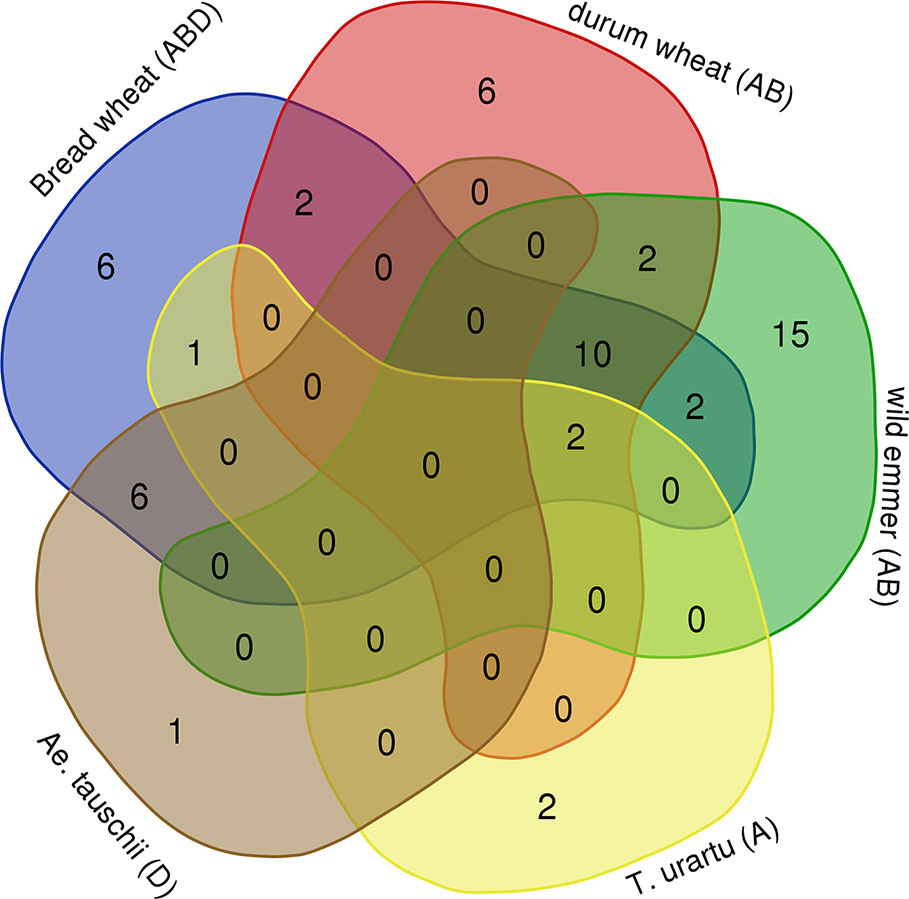
Figure 2 Common and unique Mariam insertions. Venn diagram shows the amount of common and unique insertions to each species. 2 insertions of A sub-genome were common to T. urartu, wild emmer, durum and bread wheat, 6 insertions of D sub-genome were common to Ae. tauschii and bread wheat, 10 insertions of A or B sub-genomes were common to durum, wild emmer and bread wheat. Ae. tauschii had only one unique insertion that was not found in bread wheat, T. urartu had 2 unique insertions, durum and bread wheat each had its own 6 unique insertions, while wild emmer had 15 unique insertions (not found in any other species). The diagram was generated using tools of Ghent University (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Mariam2—A Second Variant of Mariam Family
The computer-assisted analysis of Mariam insertions led to the discovery of a second variant of Mariam in wheat termed Mariam2 (291bp), suggesting this family has been divided over time into two sub-families. To validate the results and exclude sequence errors, we retrieved sequences using the second version of Mariam as a query sequence using MAK software and BLASTN analysis from wheat genome drafts. To this end, 34 Mariam2 insertions were retrieved from wheat genome drafts: 10 insertions were found in wild emmer wheat, 9 in bread wheat, 10 in durum, 3 in Ae. tauschii and 2 in T. urartu. A consensus sequence of Mariam2 was built based on MSA analysis of the retrieved sequences (Supplementary Figure 1). In addition, relatively high conservation (over 90% sequence similarity) among Mariam2 full-length insertions was observed (Supplementary Figure 2). Analysis of target site duplication (TSD) showed that Mariam2 full-length insertions possess a 9 bp (GTTACAAAC) TSD (Supplementary Figure 3). Alignment of the two consensus sequences of Mariam1 and Mariam2 (using MAFFT software, Supplementary Figure 1) showed high sequence similarity in the first 39 bp and the last 50 bp, and two relatively large gaps; one at ~111-132 bp of the alignment suggesting deletion in Mariam2 variant and a second gap at ~168-186 bp of the alignment suggesting deletion in Mariam1 variant. To examine the similarity of Mariam2 sequences to Mariam1, a phylogenetic tree analysis by maximum likelihood was performed for all full-length sequences. The phylogenetic tree has divided insertions into 2 major groups (Figure 3), the first group consists mainly of Mariam1 insertions, and two Mariam2 insertions (D7-1 and D6-2) that showed partial similarity to insertions of Mariam1 (D4-2, D4-4, A4-5, A3-1 and B7-4), indicating these sequences might be intermediate between the two versions of Mariam. The second group was separated into sub-clusters, one consists of Mariam2 insertions (100% bootstrap repetitions) and the other sub-clusters consist of Mariam1 insertions. A phylogenetic tree based on Multiple sequence alignment of all Mariam2 insertions, showed separation into 2 major groups, while insertions were clustered to smaller groups mostly by a genome-specific manner (Figure 4).
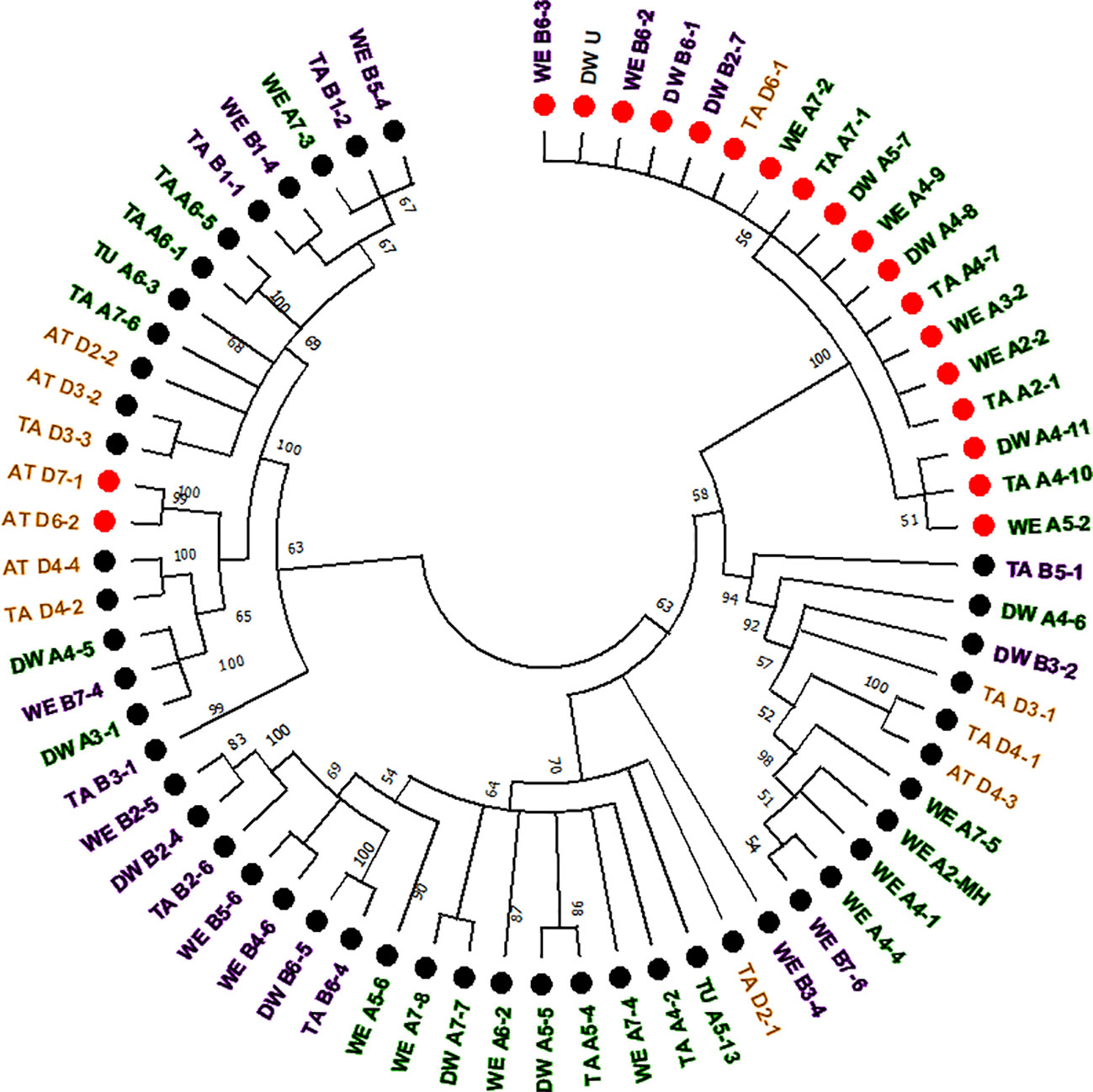
Figure 3 Phylogenetic tree of full-length sequences of Mariam1 and Mariam2. Insertion codes are composed of the species (TA—bread wheat, AT—Ae. tauschii, TU—T. urartu, WE—wild emmer and DW—durum wheat) sub-genome (A/B/D), chromosome number (1-7\U) and a serial number. Black dots represent Mariam1 insertions and red dots represent Mariam2 insertions. Insertions from A sub-genome are marked with green, insertions of B sub-genome are marked with purple, insertions of D sub-genome are marked with orange and insertions of U (unknown) were marked as black. The insertions were clustered into 2 major groups, one containing mostly Mariam1 insertions and two Mariam2 insertions (D7-1 and D6-2). The second group was separated to smaller clusters, one containing only Mariam2 insertions and the other clusters contained only Mariam1 insertions. A 45% cutoff parameter was used in this tree. The percentage of replicate trees in which the insertions clustered together in the bootstrap test (100 replicates) are shown next to the branches.
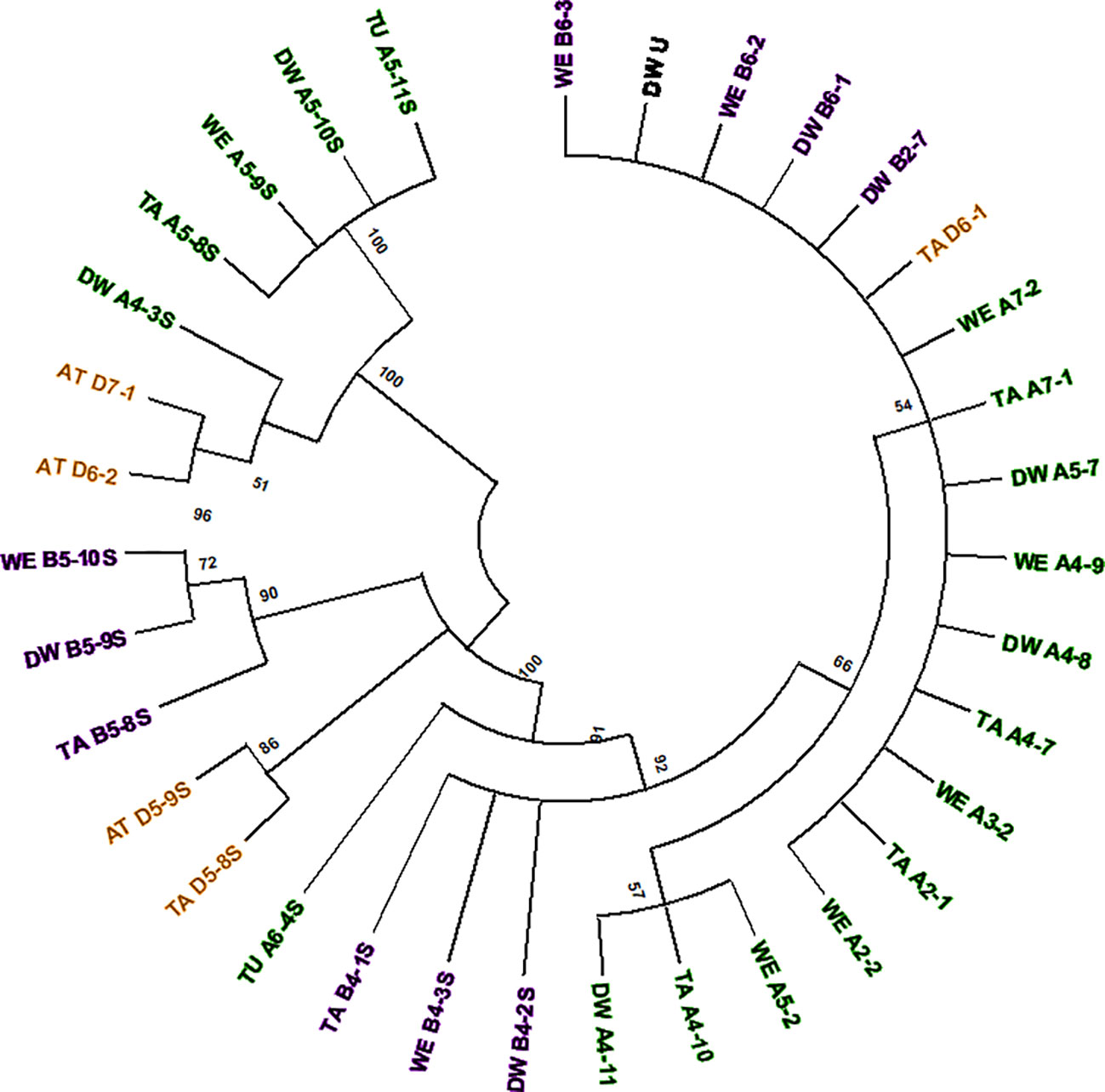
Figure 4 Phylogenetic tree of all Mariam2 insertions. Insertion codes are composed of the species (TA—bread wheat, AT—Ae. tauschii, TU—T. urartu, WE—wild emmer, and DW—durum wheat) sub-genome (A/B/D), chromosome number (1-7\U) and a serial number. Insertions from A sub-genome are marked with green, insertions of B sub-genome are marked with purple and insertions of D sub-genome are marked with orange. The phylogenetic tree showed separation to two groups, while insertions were mostly clustered according to their sub-genome (A/B/D) and synthetic loci of different species. A 45% cutoff parameter was used in this tree. The percentage of replicate trees in which the insertions clustered together in the bootstrap test (100 replicates) are shown next to the branches.
Analysis of common insertions of Mariam2 in wheat species according to flanking sequences, showed one short insertion that was found within intron 1 of a gene coding for MUSE14, a TRAF domain protein (gene acc. TRIUR3_28945 and TRIDC5AG009460) and was conserved among chromosome 5 of all different wheat species and sub-genomes of wheat. Site-specific PCR validation showed a conserved insertion site in diploid and polyploid wheat accessions (Supplementary Figure 4A). In two other cases, the insertions found were common to the allopolyploid species (durum, wild emmer and bread wheat)—A4-7/8/9 and B4-1/2/3S (Figure 4). In another two cases, the insertions were common to Ae. tauschii and bread wheat– D5-8/9S and D6-1/2 (Figure 4), suggesting these are relatively old elements that were inherited from Ae. tauschii to bread wheat.
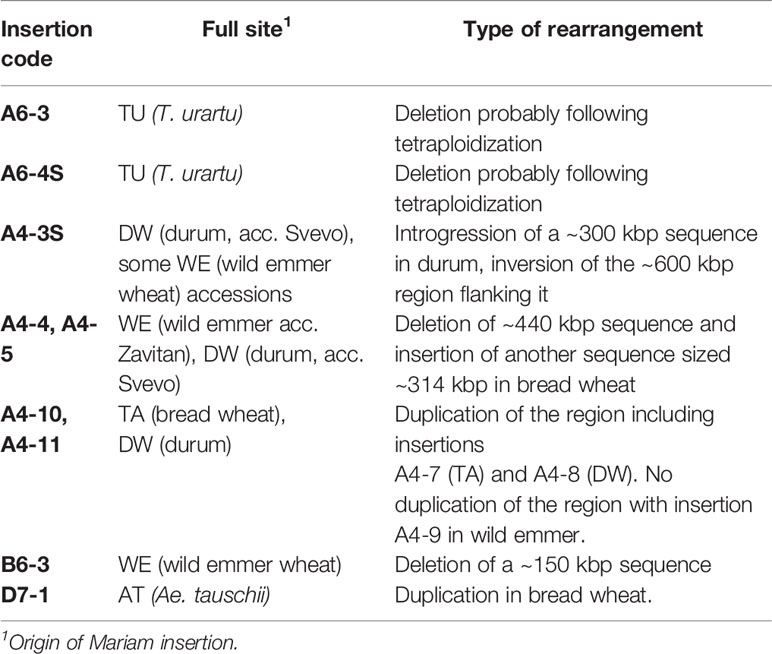
Genomic Rearrangement Events of Mariam-Containing Sequences in Wheat
In the computer-assisted analysis we assessed events of genomic rearrangements in wheat, including large INDELs (deletions/insertions) and sequence introgressions (Table 1). We have assessed cases of INDELs of Mariam-containing sequences. For example, two Mariam insertions (A6-3, Figure 3; and A6-4S, Figure 4) that were found in T. urartu were absent in the allopolyploid genome drafts. Site-specific PCR analysis with primers flanking the insertions showed bands amplified only in T. urartu accessions (Supplementary Figures 4B, C), indicating that insertion sites of A6-3 and A6-4S underwent deletion in the allopolyploid species.
Insertion A4-3S (Figure 5) was unique to chromosome 4A in durum (coordinates 4A: 619336977-619336798) and absent in other wheat genome drafts (Table 1). A comparison of a larger portion of chromosome 4A of durum (Svevo) vs. chromosome 4A of wild emmer (Zavitan) and bread wheat (Chinese Spring), suggested that Mariam insertion is located within a region that was involved in a possible introgression event of a 289,185 bp sequence in durum (coordinates 4A: 619,072,836- 619,362,021). Further analysis of the syntenic locus in wild emmer and bread wheat drafts showed that the downstream sequence of the introgression underwent an inversion event of ~600 kbp (coordinates in durum 4A: 619,362,021- 619,955,471), while the upstream sequence (~400 kbp) of the introgression was conserved (Figure 5, see also dot plot analysis in Supplementary Figure 5A). Upstream to the ~600 kbp inversion, an insertion of 140 kbp was found in durum (Svevo), while a different sequence of ~40 kbp was found at the same site in wild emmer (Zavitan) and bread wheat (Chinese Spring). Site specific PCR validation (Supplementary Figure 4D) for the flanking sequences of insertion A4-3S showed a full site in durum (Svevo), Ae. tauschii (acc. 603243 from Pakistan), and 3 accessions of wild emmer (TTD160-Syria, A1 and A23 (Amiad, Israel)). Note that all bands were extracted from the gel and sequenced for validation.
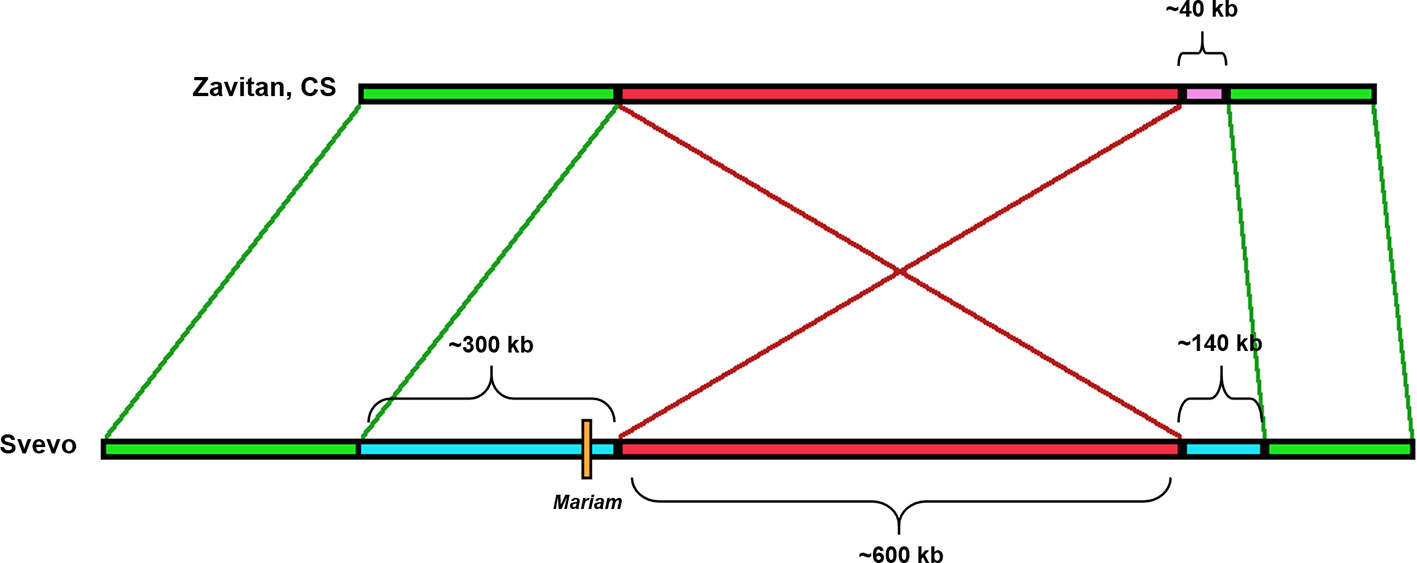
Figure 5 Schematic diagram of the rearrangement in which insertion A4-3S was involved in. Mariam (orange box) was part of a large insertion (blue box) of ~300 kbp in Svevo (DW). One side flanking this insertion showed an inversion (red box) of 600 kbp, and the other showed a sequence of ~400 kbp that was conserved in Zavitan (WE) and bread wheat (CS) as well. Upstream to the ~600 kbp inversion, an insertion of 140 kbp was found in Svevo, while a different sequence of ~40 kbp was found at the same site in Zavitan and CS. The green boxes represent conserved sites.
Molecular characterization of the locus that underwent introgression showed that this sequence includes 4 disease resistance genes (TRITD4Av1G217440, TRITD4Av1G217450, TRITD4Av1G217590, TRITD4Av1G217600) as well as Anthocyanin 5-aromatic acyltransferase (TRITD4Av1G217510) coding gene. In addition, it is part of some QTLs that are associated with biomass (QTL0782_BM-Mengistu_et_al:2016, QTL0789_BM-Mengistu_et_al:2016, QTL0788_BM-Mengistu_et_al:2016), spikelets per spike (QTL1921_4A-Roncallo_et_al._2018), height (QTL1920_4A-Roncallo_et_al._2018), spike density (QTL1062_4A-Distelfeld_et_al.) and more (see https://wheat.pw.usda.gov/jb/?data=/ggds/whe-svevo2018).
One insertion was common to durum (Svevo, A4-5) and wild emmer (Zavitan, A4-4, Figure 1) but was not found in bread wheat and Ae. tauschii (Table 1). Site specific PCR validation (Supplementary Figure 4E) with primers flanking this insertion showed full site in durum and in different accessions of wild emmer; Zavitan (Israel), TTD32 (Turkey), A16b (Amiad, Israel), J1 (Jaba, Israel), T8, T1 and T2 (Tabja, Israel). Analysis of this locus in bread wheat genome draft revealed deletion of a ~440 kbp sequence that included this insertion (see dot plot in Supplementary Figure 5B). The deleted sequence contains mainly LTR retrotransposons, and CACTA transposons. In addition, a unique ~ 314 kbp insertion was found in bread wheat at the syntenic locus (coordinates: 688,952,833-689,266,972).
Insertions A4-10 and A4-11 (Figure 4) were found within a duplicated locus of the sequence that contains insertions A4-7 and A4-8 (Figure 4) in durum and bread wheat (Table 1). However, no duplication was detected in wild emmer, when the locus that contains A4-9 (Figure 4) insertion was tested.
Insertion B6-3 (Figure 4) was found in-silico only in wild emmer. Molecular characterization of the insertion site locus showed that a sequence of ~150 kbp, including the insertion in wild emmer (coordinates 6B: 499,153,784 - 499,306,375), was absent in durum and bread wheat genome drafts (Table 1, see dot plot results in Supplementary Figure 5C). Site-specific PCR validation using primers flanking B6-3 insertion revealed that this insertion is found in most of wild emmer wheat accessions (Supplementary Figure 4F) and in one accession of bread wheat (acc. 129526).
Insertion D7-1 (Figure 4) was mapped to chromosome 7 of Ae. tauschii and was not found in bread wheat genome draft. A BLASTN analysis of the regions flanking D7-1 insertion showed a ~830 bp duplication of the sequence in chromosome D7 of bread wheat (coordinates, D7:32026174-32026998 and D7: 35166235-35167066) (Table 1).
Discussion
The discovery of Mariam, a wheat-unique miniature transposable element family, was first reported in our previous study (Domb et al., 2019). We have suggested this family can be classified into Mutator superfamily of MITEs (Miniature Inverted-repeat Transposable Elements) due to the 9 bp varying TSD (target site duplication) found in some insertions and the lack of TIRs (Lisch, 2002; Wicker et al., 2007). In this study, we have focused on the dynamics of Mariam family in different wheat species, using the genome drafts of T. urartu (TU, donor of A genome), Ae. tauschii (AT, donor of D genome), durum wheat (DW, T. turgidum ssp. durum, tetraploid of AB genome), wild emmer (WE, T. turgidum ssp. diccocoides, tetraploid AB genome) and bread wheat (TA, T. aestivum, hexaploidy ABD genome), as well as the genetic material of different wheat accessions.
Retrieval of Mariam elements from wheat genome drafts revealed different insertion lengths, ranging between 93 up to 337 bp. During the in-silico analysis of Mariam insertions, we have discovered two different variants with similar ends but a relatively different internal part, suggesting there are two sub-families of Mariam (Figure 3 and Supplementary Figure 1). We have retrieved a total of 94 Mariam insertions from all 5 wheat genome drafts, of these, 60 were of sub-family Mariam1 and 34 insertions were of sub-family Mariam2. A comparison of the flanking sequences of Mariam insertions has indicated the proportions of conserved vs. unique insertions, meaning ancient insertions inherited from progenitors or relatively new insertions indicating later activity. Wild emmer wheat presented the highest number of unique insertions suggesting that Mariam was probably recently active in wild emmer, similarly to our previous report on Mariam1 based on intra-specific insertional-polymorphism analysis (Domb et al., 2019).
One of the major questions regarding the mobility of plant transposable elements is whether they transpose only within proximate regions on the same chromosome (“local hopping”) or between different chromosomes as shown in mammals (Kim, 2009; Liang et al., 2009) and even between different sub-genomes. Our analysis of Mariam insertions in wild emmer and bread wheat showed clusters of insertions from different chromosomes and from different sub-genomes. In some cases, clustering of insertions from homeologs chromosomes of different sub-genomes might indicate an ancient insertion in syntenic loci that has transposed in the progenitor of the diploid wheat species. In this analysis, we have found clustering of non-homeolog chromosomes from different sub-genomes, indicating a possible transposition between different chromosomes and even between different sub-genomes.
Miniature TEs are considered to be very abundant in genic regions and in some cases act as regulators of gene expression (Rebollo et al., 2012; Vaschetto, 2016; Wicker et al., 2018). Insertions into coding sequences usually disrupt gene function while insertions into introns, promotors or near genes can alter its expression or splicing (Dubin et al., 2018). Some Mariam insertions were found within or close to protein coding genes. For example, one insertion (A2-MH) was discussed in our previous study (Domb et al., 2019) where we showed this insertion into the CDS of a gene coding for 5-formyltetrahydrofolate cyclo-ligase leads to disruption of the ORF by insertion of a premature stop codon, that leads if translated to shorter with altered C-terminus protein. Another insertion, discussed in this study, was found within an intron 1 of a gene coding for MUSE14 and was found to be very ancient and conserved in all species and all sub-genomes suggesting that transposition occurred probably in the progenitor of diploid wheat species (over ~4 million years ago) and had a neutral or beneficial effect, as it has been fixed (domesticated) during the evolution of wheat.
Analysis of polymorphic insertions showed some cases of conserved loci with empty sites (excision/no insertion), some cases where only one side of the flanking region of Mariam was found and other cases in which the syntenic region was not found in other wheat species, suggesting a rearrangement has occurred at these sites (Table 1). We have analyzed cases of rearrangements that involved Mariam elements and found cases of large deletions, INDELs (deletions/insertions) and sequence introgressions in polyploid wheat species. It is important to mention that although the most updated genome drafts of Aegilops and Triticum were used, errors might occur due to incomplete sequence assembly. For this reason, wet-bench validation is required to assess the integrity of the results. In this study, we have validated using site-specific PCR analysis a subset of cases that were further analyzed (Supplementary Figure 4).
Transposable elements can be good evolutionary markers to study phylogenetics as well as genetic diversity among populations (Tatout et al., 1999; Queen et al., 2004; Kalendar et al., 2011; Yaakov et al., 2012; Tagimanova et al., 2015; Domb et al., 2017). Some transposable elements can be used as markers for the discovery of large-scale chromosomal rearrangements (Devos et al., 2002; Bennetzen, 2005; Kraitshtein et al., 2010; Yaakov et al., 2012; Bariah et al., 2020) and for crop improvement (Venkatesh and Nandini, 2020). Although Mariam is a low copy number family, its dynamics throughout the evolutionary history of wheat are quite interesting and its insertional polymorphism can be used to discover cases of large-scale rearrangements. Mariam was found to be a good marker for genetic diversity in populations of wild emmer wheat (Domb et al., 2019), for polymorphism in wheat species and discovery of rearrangements. With the advancement of sequencing technologies, the use of MITEs as markers in different bioinformatic analyses is becoming more and more relevant.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
DKF – design of the study, experiments, analyses and manuscript writing. IB –analyses of rearrangements and figures preparation, KD – design of the study and experiments, KK - design of the study and manuscript writing. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by Israel Science Foundation (grant 322/15) to KK. The authors declare that ISF is the only funding source of this work.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Vadim khasdan and Alon Ziv for their kind assistance in the lab.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01173/full#supplementary-material
References
Appels, R., Eversole, K., Feuillet, C., Keller, B., Rogers, J., Stein, N., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, 661–673. doi: 10.1126/science.aar7191
Avni, R., Nave, M., Barad, O., Baruch, K., Twardziok, S. O., Gundlach, H., et al. (2017). Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357, 93–97. doi: 10.1126/science.aan0032
Bao, W., Kojima, K. K., Kohany, O. (2015). Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11. doi: 10.1186/s13100-015-0041-9
Bariah, I., Keidar-Friedman, D., Kashkush, K. (2020). Identification of large-scale genomic rearrangements during wheat evolution and the underlying mechanisms. PloS One 15, e0231323. doi: 10.1371/journal.pone.0231323
Bennetzen, J. L. (2005). Transposable elements, gene creation and genome rearrangement in flowering plants. Curr. Opin. Genet. Dev. 15, 621–627. doi: 10.1016/j.gde.2005.09.010
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: Architecture and applications. BMC Bioinf. 10, 421. doi: 10.1186/1471-2105-10-421
Capy, P., Gasperi, G., Biémont, C., Bazin, C. (2000). Stress and transposable elements: Co-evolution or useful parasites? Heredity (Edinb). 85, 101–106. doi: 10.1046/j.1365-2540.2000.00751.x
Casacuberta, J. M., Santiago, N. (2003). Plant LTR-retrotransposons and MITEs: Control of transposition and impact on the evolution of plant genes and genomes. Gene 311, 1–11. doi: 10.1016/S0378-1119(03)00557-2
Charles, M., Belcram, H., Just, J., Huneau, C., Viollet, A., Couloux, A., et al. (2008). Dynamics and differential proliferation of transposable elements during the evolution of the B and A genomes of wheat. Genetics 180, 1071–1086. doi: 10.1534/genetics.108.092304
Clavijo, B. J., Venturini, L., Schudoma, C., Accinelli, G. G., Kaithakottil, G., Wright, J., et al. (2017). An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res. 27, 885–896. doi: 10.1101/gr.217117.116
Devos, K. M., Brown, J. K. M. M., Bennetzen, J. L. (2002). Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12, 1075–1079. doi: 10.1101/gr.132102
Domb, K., Keidar, D., Yaakov, B., Khasdan, V., Kashkush, K. (2017). Transposable elements generate population-specific insertional patterns and allelic variation in genes of wild emmer wheat (Triticum turgidum ssp. dicoccoides). BMC Plant Biol. 17, 175. doi: 10.1186/s12870-017-1134-z
Domb, K., Keidar-Friedman, D., Kashkush, K. (2019). A novel miniature transposon-like element discovered in the coding sequence of a gene that encodes for 5-formyltetrahydrofolate in wheat. BMC Plant Biol. 19, 1–11. doi: 10.1186/s12870-019-2034-1
Dubin, M. J., Mittelsten Scheid, O., Becker, C. (2018). Transposons: a blessing curse. Curr. Opin. Plant Biol. 42, 23–29. doi: 10.1016/j.pbi.2018.01.003
Feldman, M., Levy, A. A. (2005). Allopolyploidy - A shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 109, 250–258. doi: 10.1159/000082407
Gray, Y. H. M., Genetics, M., Group, E., Sciences, B., National, A. (2000). It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet. 16, 461–468. doi: 10.1016/S0168-9525(00)02104-1
Jiang, Y. F., Chen, Q., Wang, Y., Guo, Z. R., Xu, B. J., Zhu, J., et al. (2019). Re-acquisition of the brittle rachis trait via a transposon insertion in domestication gene Q during wheat de-domestication. New Phytol. 224, 961–973. doi: 10.1111/nph.15977
Kalendar, R., Flavell, A. J., Ellis, T. H. N. N., Sjakste, T., Moisy, C., Schulman, A. H. (2011). Analysis of plant diversity with retrotransposon-based molecular markers. Heredity (Edinb). 106, 520–530. doi: 10.1038/hdy.2010.93
Kashkush, K., Feldman, M., Levy, A. A. (2003). Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33, 102–106. doi: 10.1038/ng1063
Keidar, D., Doron, C., Kashkush, K. (2018). Genome-wide analysis of a recently active retrotransposon, Au SINE, in wheat: content, distribution within subgenomes and chromosomes, and gene associations. Plant Cell Rep. 37, 193–208. doi: 10.1007/s00299-017-2213-1
Keidar-Friedman, D., Bariah, I., Kashkush, K. (2018). Genome-wide analyses of miniature inverted-repeat transposable elements reveals new insights into the evolution of the triticum-Aegilops group. PloS One 13, e0204972. doi: 10.1371/journal.pone.0204972
Kidwell, M. G. (2002). Transposable elements and the evolution of genome size in eukaryotes. Genetica 115, 49–63. doi: 10.1023/A:1016072014259
Kim, I. (2009). A Transposon-Based Genetic Screen in Mice Identifies Genes Altered in Colorectal Cancer. Science 323, 1747–1751. doi: 10.1126/science.1163040
Kraitshtein, Z., Yaakov, B., Khasdan, V., Kashkush, K. (2010). Genetic and epigenetic dynamics of a retrotransposon after allopolyploidization of wheat. Genetics 186, 801–812. doi: 10.1534/genetics.110.120790
Krull, M., Brosius, J., Schmitz, J. (2005). Alu-SINE exonization: En route to protein-coding function. Mol. Biol. Evol. 22, 1702–1711. doi: 10.1093/molbev/msi164
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lev-Maor, G., Sorek, R., Shomron, N., Ast, G. (2003). The birth of an alternatively spliced exon: 3′ Splice-site selection in Alu exons. Science 300, 1288–1291. doi: 10.1126/science.1082588
Levy, A. A., Feldman, M. (2002). The impact of polyploidy on grass genome evolution. Plant Physiol. 130, 1587–1593. doi: 10.1104/pp.015727
Levy, A. A., Feldman, M. (2004). Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization. Biol. J. Linn. Soc 82, 607–613. doi: 10.1111/j.1095-8312.2004.00346.x
Li, J., Wang, Z., Peng, H., Liu, Z. (2014). A MITE insertion into the 3′-UTR regulates the transcription of TaHSP16.9 in common wheat. Crop J. 2, 381–387. doi: 10.1016/j.cj.2014.07.001
Liang, Q., Kong, J., Stalker, J., Bradley, A. (2009). Chromosomal mobilization and reintegration of Sleeping Beauty and PiggyBac transposons. Genesis 47, 404–408. doi: 10.1002/dvg.20508
Ling, H.-Q. Q., Ma, B., Shi, X., Liu, H., Dong, L., Sun, H., et al. (2018). Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature 557, 424–428. doi: 10.1038/s41586-018-0108-0
Lisch, D. (2002). Mutator transposons. Trends Plant Sci. 7, 498–504. doi: 10.1016/S1360-1385(02)02347-6
Lisch, D. (2013). How important are transposons for plant evolution? Nat. Rev. Genet. 14, 49–61. doi: 10.1038/nrg3374
Luo, M. C., Gu, Y. Q., Puiu, D., Wang, H., Twardziok, S. O., Deal, K. R., et al. (2017). Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551, 498–502. doi: 10.1038/nature24486
Maccaferri, M., Harris, N. S., Twardziok, S. O., Pasam, R. K., Gundlach, H., Spannagl, M., et al. (2019). Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 51, 885–895. doi: 10.1038/s41588-019-0381-3
Mansour, A. (2007). Epigenetic activation of genomic retrotransposons. J. Cell Mol. Biol. 6, 99–107.
Middleton, C. P., Stein, N., Keller, B., Kilian, B., Wicker, T. (2013). Comparative analysis of genome composition in Triticeae reveals strong variation in transposable element dynamics and nucleotide diversity. Plant J. 73, 347–356. doi: 10.1111/tpj.12048
Nevo, E., Beiles, A. (1989). Genetic diversity of wild emmer wheat in Israel and Turkey. Theor. Appl. Genet. 77, 421–455. doi: 10.1007/BF00305839
Okonechnikov, K., Golosova, O., Fursov, M., Varlamov, A., Vaskin, Y., Efremov, I., et al. (2012). Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 28, 1166–1167. doi: 10.1093/bioinformatics/bts091
Petersen, G., Seberg, O., Yde, M., Berthelsen, K. (2006). Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum). Mol. Phylogenet. Evol. 39, 70–82. doi: 10.1016/j.ympev.2006.01.023
Queen, R. A., Gribbon, B. M., James, C., Jack, P., Flavell, A. J. (2004). Retrotransposon-based molecular markers for linkage and genetic diversity analysis in wheat. Mol. Genet. Genomics 271, 91–97. doi: 10.1007/s00438-003-0960-x
Rebollo, R., Romanish, M. T., Mager, D. L. (2012). Transposable Elements: An Abundant and Natural Source of Regulatory Sequences for Host Genes. Annu. Rev. Genet. 46, 21–42. doi: 10.1146/annurev-genet-110711-155621
Sabot, F., Simon, D., Bernard, M. (2004). Plant transposable elements, with an emphasis on grass species. Euphytica 139, 227–247. doi: 10.1007/s10681-004-3179-y
Schmitz, J., Brosius, J. (2011). Exonization of transposed elements: A challenge and opportunity for evolution. Biochimie 93, 1928–1934. doi: 10.1016/j.biochi.2011.07.014
Schrader, L., Schmitz, J. (2018). The impact of transposable elements in adaptive evolution. Mol. Ecol. 28, 1537–1549. doi: 10.1111/mec.14794
Slotkin, R. K., Martienssen, R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285. doi: 10.1038/nrg2072
Tagimanova, D. S., Novakovskaya, A. P., Uvashov, A. O., Khapilina, O. N., Kalendar, R. N. (2015). Use of Retrotransposon Markers for Analysing the Genetic Diversity of Wild Emmer Wheat (Triticum Dicoccoides). Biotechnol. Theory Pract. 4, 28–37. doi: 10.11134/btp.4.2015.4
Tatout, C. C., Warwick, S., Lenoir, A., Deragon, J.-M. M. (1999). Sine insertions as clade markers for wild crucifer species. Mol. Biol. Evol. 16, 1614–1621. doi: 10.1093/oxfordjournals.molbev.a026074
Vaschetto, L. M. (2016). Miniature Inverted-repeat Transposable Elements (MITEs) and their effects on the regulation of major genes in cereal grass genomes. Mol. Breed. 36, 30. doi: 10.1007/s11032-016-0440-8
Venkatesh, Nandini, B. (2020). Miniature inverted-repeat transposable elements (MITEs), derived insertional polymorphism as a tool of marker systems for molecular plant breeding. Mol. Biol. Rep. 47, 3155–3167. doi: 10.1007/s11033-020-05365-y
Venske, E., Dos Santos, R. S., Busanello, C., Gustafson, P., Costa de Oliveira, A. (2019). Bread wheat: a role model for plant domestication and breeding. Hereditas 156, 16. doi: 10.1186/s41065-019-0093-9
Volis, S., Song, M., Zhang, Y.-H. H., Shulgina, I. (2014). Fine-Scale Spatial Genetic Structure in Emmer Wheat and the Role of Population Range Position. Evol. Biol. 41, 166–173. doi: 10.1007/s11692-013-9256-1
Wicker, T., Matthews, D. E., Keller, B. (2002). TREP: A database for Triticeae repetitive elements. Trends Plant Sci. 7, 561–562. doi: 10.1016/S1360-1385(02)02372-5
Wicker, T., Sabot, F., Hua-Van, A., Bennetzen, J. L., Capy, P., Chalhoub, B., et al. (2007). A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8, 973–982. doi: 10.1038/nrg2165
Wicker, T., Gundlach, H., Spannagl, M., Uauy, C., Borrill, P., Ramirez-Gonzalez, R. H., et al. (2018). Impact of transposable elements on genome structure and evolution in bread wheat. Genome Biol. 19, 103. doi: 10.1186/s13059-018-1479-0
Yaakov, B., Ceylan, E., Domb, K., Kashkush, K. (2012). Marker utility of miniature inverted-repeat transposable elements for wheat biodiversity and evolution. Theor. Appl. Genet. 124, 1365–1373. doi: 10.1007/s00122-012-1793-y
Keywords: transposable elements, Mariam, genome evolution, allopolyploidy, wheat
Citation: Keidar-Friedman D, Bariah I, Domb K and Kashkush K (2020) The Evolutionary Dynamics of a Novel Miniature Transposable Element in the Wheat Genome. Front. Plant Sci. 11:1173. doi: 10.3389/fpls.2020.01173
Received: 24 April 2020; Accepted: 20 July 2020;
Published: 31 July 2020.
Edited by:
Ruslan Kalendar, University of Helsinki, FinlandReviewed by:
Wilfried A. Kues, Friedrich Loeffler Institute (FLI), GermanyDariusz Grzebelus, University of Agriculture in Krakow, Poland
Copyright © 2020 Keidar-Friedman, Bariah, Domb and Kashkush. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Khalil Kashkush, kashkush@bgu.ac.il
†Present address: Katherine Domb, Center for Plant Molecular Biology, University of Tübingen, Tübingen, Germany
 Danielle Keidar-Friedman
Danielle Keidar-Friedman Inbar Bariah
Inbar Bariah Katherine Domb†
Katherine Domb† Khalil Kashkush
Khalil Kashkush