- Institute of Animal Genetics and Breeding, College of Animal Science and Technology, Sichuan Agricultural University, Chengdu, China
High-altitude acclimatization is a representative example of vertebrates’ acclimatization to harsh and extreme environments. Previous studies reported sufficient evidence for a molecular genetic basis of high-altitude acclimatization, and genomic patterns of genetic variation among populations and species have been widely elucidated in recent years. However, understanding of the miRNA role in high-altitude acclimatization have lagged behind, especially in non-model species. To investigate miRNA expression alterations of goats that were induced by high-altitude stress, we performed comparative miRNA transcriptome analysis on six hypoxia-sensitive tissues (heart, kidney, liver, lung, skeletal muscle, and spleen) in two goat populations from distinct altitudes (600 and 3000 m). We obtained the expression value of 1391 mature miRNAs and identified 138 differentially expressed (DE) miRNAs between high and low altitudes. Combined with tissue specificity analysis, we illustrated alterations of expression levels among altitudes and tissues, and found that there were coexisting tissue-specific and -conserved mechanisms for hypoxia acclimatization. Notably, the interplay between DE miRNA and DE target genes strongly indicated post-transcriptional regulation in the hypoxia inducible factor 1, insulin, and p53 signaling pathways, which might play significant roles in high-altitude acclimatization in domestic goats. It’s also worth noting that we experimentally confirmed miR-106a-5p to have a negative regulation effect on angiogenesis by directly targeting FLT-1. These results provide insight into the complicated miRNA expression patterns and regulatory mechanisms of high-altitude acclimatization in domestic goats.
Introduction
Environmental changes impose the pressure of natural selection on vertebrates, which supports the evolution of adaptive mechanisms for survival. As a typical example of adaptive evolution in extreme environments (Richalet, 2007), high-altitude acclimatization refers to the heritable and irreversible changes in morphology (Brutsaert et al., 1999; Miles et al., 2009), physiology (Storz et al., 2009; Naeije, 2010), biochemistry (García-Hjarles, 1989; Li et al., 2010), and ethology (Mamatov et al., 2012) in highland habitants during long-term selection pressure, such as reduced oxygen availability, low ambient temperatures, and high ultraviolet radiation (UV; Brookfield and Allen, 1989; Wang et al., 2014). These changes are primarily triggered on a molecular level to modulate organic metabolism, especially in representatively hypoxia-sensitive tissues such as heart (Jarmakani et al., 1978; Wilson et al., 1993; Moromisato et al., 1996; Park et al., 2007; Chen et al., 2009), kidney (Luks et al., 2008; Dan et al., 2010; Yijiang et al., 2013), liver (Komolova and Egorov, 1985; Moromisato et al., 1996; Rong and Gastroenterology, 2009), lung (Moromisato et al., 1996; Liu, 2011; Li et al., 2012), longissimus muscle (Lundby et al., 2003, 2009; Murray, 2009; Levett et al., 2012; Zhang et al., 2015), and spleen (Ou et al., 1980; Kam et al., 1999; Richardson et al., 2008). Previous studies reported sufficient evidence for a molecular genetic basis of high-elevation acclimatization (Breen et al., 2008; Simonson et al., 2010; Scott et al., 2011; Bigham et al., 2013; Yang et al., 2017), and genomic patterns of genetic variation among populations and species have been widely elucidated in recent years (Cheviron and Brumfield, 2012; Mingzhou et al., 2013; Qu et al., 2013; Ai et al., 2014; Zhou et al., 2014). For instance, key erythropoiesis-related genes including EPAS1, PRARA, and EGLN1 were found mutated in Tibetans, which were involved in hypoxia inducible factor 1 (HIF-1) signaling pathway and modulate erythropoiesis in a HIFs-centered manner. Independent or synergetic variants in these genes might be responsible for erythropoiesis responses to hypoxia in Tibetans (Simonson et al., 2010). Moreover, researchers found that the RGCC gene was under strong selection during high-altitude adaptation of Tibetan pig, which plays a key role in hypoxia-induced anti-angiogenesis. Interacted with HIF-1α and VEGF, preferentially selected variants in RGCC may inhibit the capillary growth and consequently avoid capillary leak in Tibetan pigs under long-term exposure to the hypoxic environment (Ai et al., 2014).
microRNAs (miRNAs) are a class of endogenous ∼20-nt non-coding RNAs that regulate gene expression at the post-transcriptional level via imperfect matches in their target mRNA 3’UTR regions and consequential translational repression or mRNA degradation (Bartel, 2004, 2009). Recent research on human and model animals revealed that miRNAs serve as important regulators in biological processes implicated in acclimatizations to hypoxia (Pocock, 2011), cold exposure (Al-Fageeh and Smales, 2006; Pilotte et al., 2011), and UV damage (Wan et al., 2011), which are environmental conditions associated with high altitudes (Bigham et al., 2013). For example, the miR-15 family is regulated by hypoxia and predicted to influence cardiomyocyte cell survival by regulating the expression of several pro-survival proteins (Hitoo et al., 2010; Lin et al., 2010; Small et al., 2010; Dejean et al., 2011). The UV-induced miR-211 regulates the UV-responsive cell-cycle control gene p27 and affects the proliferation potential of human prostate carcinoma cell lines (Silvia et al., 2007; Pothof et al., 2009). The well-documented hypoxia-regulated microRNA (HRM), miR-210, is directly regulated by HIF-1 (Huang et al., 2009) and participates in DNA damage response (Crosby et al., 2009), angiogenesis (Camps et al., 2008), apoptosis (Kulshreshtha et al., 2007), and mitochondrial respiration (Chan et al., 2009). However, understandings in the role of miRNA in high-altitude acclimatization have mainly focused on either human or model animals, and research on natural populations of non-model species has lagged behind.
While previous research on high-altitude goats mostly described their phenotypic characteristics, geographical distribution and living habits, molecular mechanisms behind their high-altitude acclimatizations have been poorly illustrated. As the genetic characteristics remain unclear, many important economic traits could not been fully utilized. One way for breakthrough is to study the adaptational changes of goats at small RNA transcriptome level. Therefore, we performed comparative miRNA transcriptome analysis on two populations of domestic goats, which are typical agricultural animals that reside in geographically neighboring regions at distinct altitudes (600 and 3000 m). Six hypoxia-sensitive tissues (heart, kidney, liver, lung, longissimus muscle, and spleen) were sampled from each individual and 36 small RNA libraries were constructed for sequencing.
Results
Data Summary
We selected three females as biological replicates from the low- and high-altitude domestic goat populations, and sampled six representative hypoxia-sensitive tissues (heart, kidney, liver, lung, longissimus muscle, and spleen) from each replicate. In total, 36 samples were separately used to construct small RNA libraries with an average RNA integrity number (RIN) of 7.97 (7.97 ± 0.79, n = 36), which were overall appropriate for small RNA-seq (Supplementary Table S1). Deep sequencing generated an average of 12.25 million (M) (12.25 ± 1.09 M, n = 36) raw reads for each library, and resulted in a total of 441.17 M raw reads. Through a series of quality control procedures, an average of 11.99 M (11.99 ± 0.93 M, n = 36) reads were retained as high-quality reads for each library, and a total of 431.51 M high-quality reads were used for miRNA identification. Consistent with the structural basis of animal miRNA, high-quality reads were mainly (79.49%) between 21 and 24-nt long, with the majority being 22-nt long (45.06%) (Supplementary Figure S1).
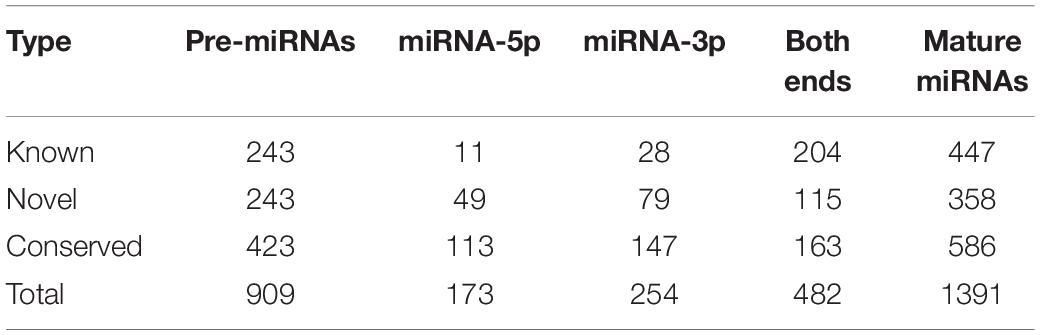
With stringent criteria in the miRDeep2 pipeline, a total of 909 pre-miRNAs that encode 1391 mature miRNAs were identified in all 36 libraries, including pre-miRNAs that were processed into either 5p or 3p mature forms, or both 5p and 3p mature forms. According to sequence conservation between identified mature miRNAs and annotated goat or other mammal miRNAs recorded in miRbase release 21 (Kozomara and Griffiths-Jones, 2014), 447 miRNAs sharing exactly the same mature sequence with goat annotated miRNAs were categorized into the “Known” group, whereas the 586 miRNAs with 100% sequence similarity to other mammalian miRNAs were categorized into the “Conserved” group, and the remainder, which did not match any annotated miRNA sequences, were categorized into the “Novel” group (Table 1).
To study the mechanisms that may underlie miRNA origins, we screened for overlapping miRNA precursor sequences in the genome with genomic elements of different types, including exons, introns of protein-coding genes and intergenic regions (Supplementary Figure S3). The analysis of genomic sources showed that, out of 909 pre-miRNAs, 426 (46.86%) were located in introns, followed by 345 pre-miRNAs (37.95%) in intergenic regions as the second largest category. Only three miRNA precursors (0.32%) were located in exons. Due to alternative splicing of protein-coding genes, 123 pre-miRNAs (13.53%) resided in both intronic and exonic regions of their host genes. A detailed analysis revealed that intronic regions were highly overrepresented with miRNA loci (permutation test, P < 0.001), which is consistent with the findings of a previous report (Berezikov, 2011).
Global miRNA Expression Profile Analysis
Through hierarchical clustering of global gene expression profiles from each sample, we found that the clustering of gene expression was tissue-dominated (Figure 1A). The samples were almost perfectly clustered by tissue and then by altitude, as directly shown by color bars above the heatmap. This expression pattern indicated that global miRNA expression varies more across tissues than between altitudes, which was consistent with the findings of a previous study (Anamaria and Henrik, 2014). Among the six tissues, the muscle and heart were more similar to each other than to other tissues, which reflects the relatively similar physiological biochemical characteristics between the two types of striated muscles (Jason et al., 2012). The clustering of biological replicates also supported experimental reliability. Results of principal components analysis (PCA) further supported these findings (Figure 1B), namely expression patterns including tissue-domination, similarity between muscle and heart, as well as the adjacent clustering of biological replicates.
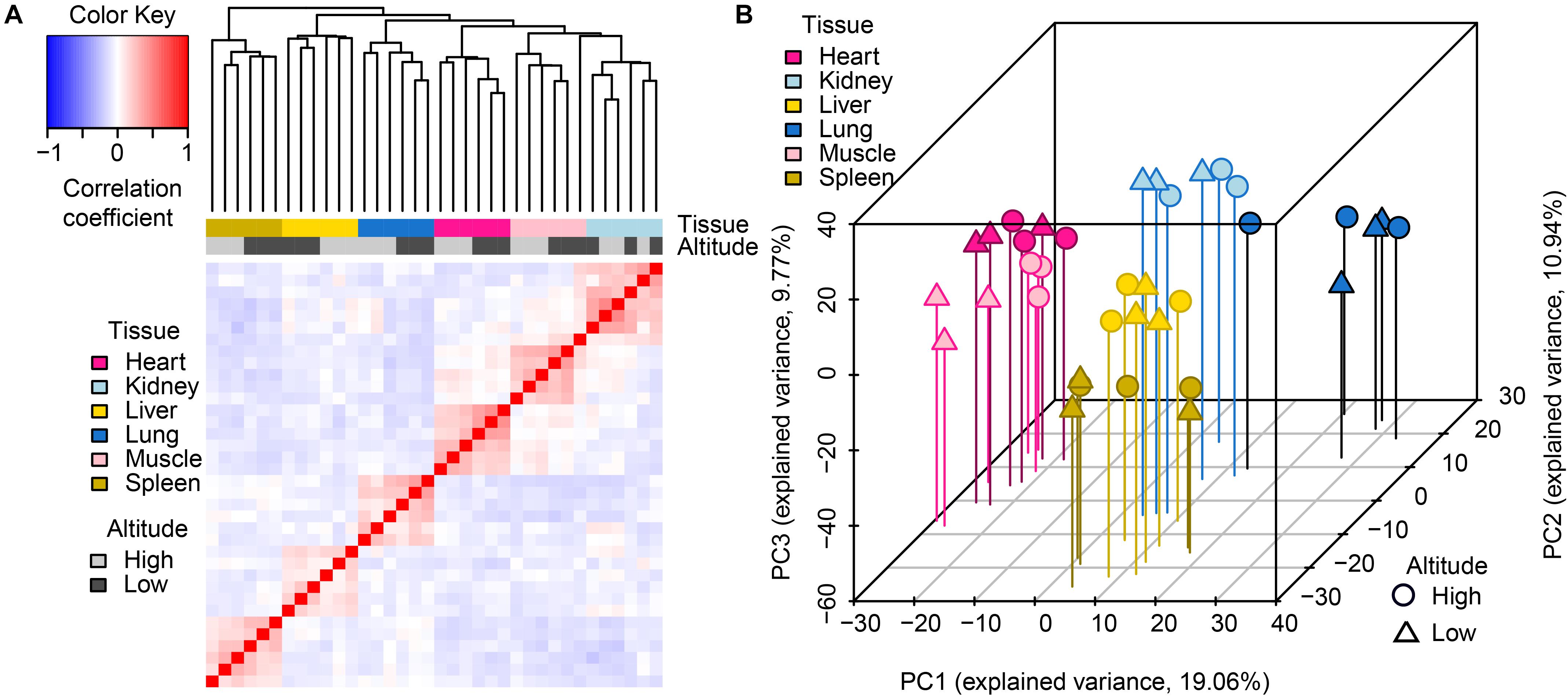
Figure 1. Global pattern of miRNA expression among tissues and altitudes. (A) Hierarchical clustering of samples using miRNA expression. Average linkage hierarchical clustering was used with the distance between samples measured by Pearson’s correlation between the vectors of z-score standardized expression values; (B) PCA of miRNA expression. The proportion of the variance explained by the principal components (PCs) is indicated in parentheses. The vertical lines with different colors from the plotted points dropping to the x/y plane show the separation of points based on the first and second PCs.
Tissue Specificity Analysis
To compare tissue specificity between miRNAs and protein-coding genes, we extracted mRNA transcriptome data sets of the corresponding goat tissues from our previous study (Tang et al., 2015), and performed tissue specific index (TSI) and tissue specific score (TSS) calculations on expression profiles of both miRNA and protein-coding genes (section “Materials and Methods”). Additionally, to test whether the tissue specificity is affected by altitudes, these calculations were separately conducted based on the expression profiles of 18 samples from high- and low-altitude goat populations (Figure 2). However, these curves indicated little impact of tissue specificity by high-altitude acclimatization on both miRNAs and protein-coding genes. A sizable proportion of miRNAs (40.47%) were tissue-specific, compared with only 12.15% of protein-coding genes, as determined by analyzing expression profiles across both altitudes.
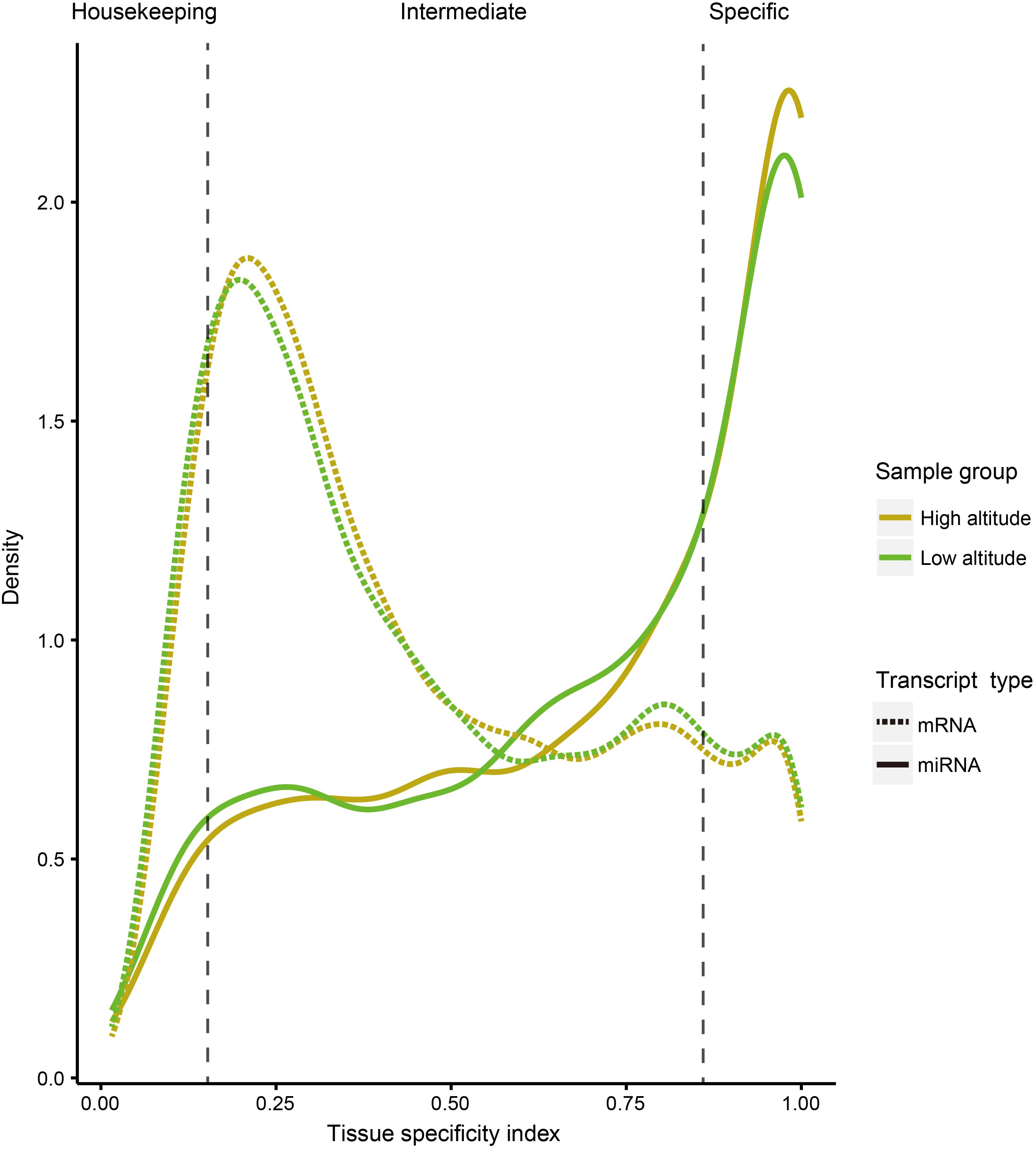
Figure 2. Distribution curve for TSI of miRNAs and protein-coding genes in different tissues. The vertical dotted lines correspond to the threshold originally proposed for defining housekeeping and specifically expressed transcripts with TSI values <0.15 and >0.85, respectively.
We then clustered tissue-specific miRNAs (Figure 3A) and genes (Figure 3B) using single-linkage clustering with expression and annotated each cluster with enriched functions of tissue-specific genes targeted by tissue-specific miRNAs within corresponding tissues (Figure 3C). Indeed, clusters of tissue-specific miRNAs and genes were enriched in functional categories specific to certain tissue. For example, target genes of the heart-specific cluster were significantly enriched in functional terms such as regulation of cardiac conduction (three genes, P = 5.89 × 10–4) and ventricular cardiac muscle cell action potential (two genes, P = 9.79 × 10–3). Muscle-specific target genes were enriched in sarcoplasmic reticulum (two genes, P = 2.03 × 10–2). Kidney-related terms, such as metanephric collecting duct development (three genes, P = 1.15 × 10–4) and sodium ion transmembrane transport (four genes, P = 6.94 × 10–4), were enriched in kidney-specific target genes. Microtubule motor activity (five genes, P = 3.02 × 10–4) and cilium-dependent cell motility (three genes, P = 4.15 × 10–2) were detected as enriched Gene Ontology (GO) terms in lung-specific genes. Additionally, over-representation of humoral immune response (three genes, P = 1.60 × 10–2) in spleen-specific genes and bile secretion (four genes, P = 6.98 × 10–4) in liver-specific genes were also found.
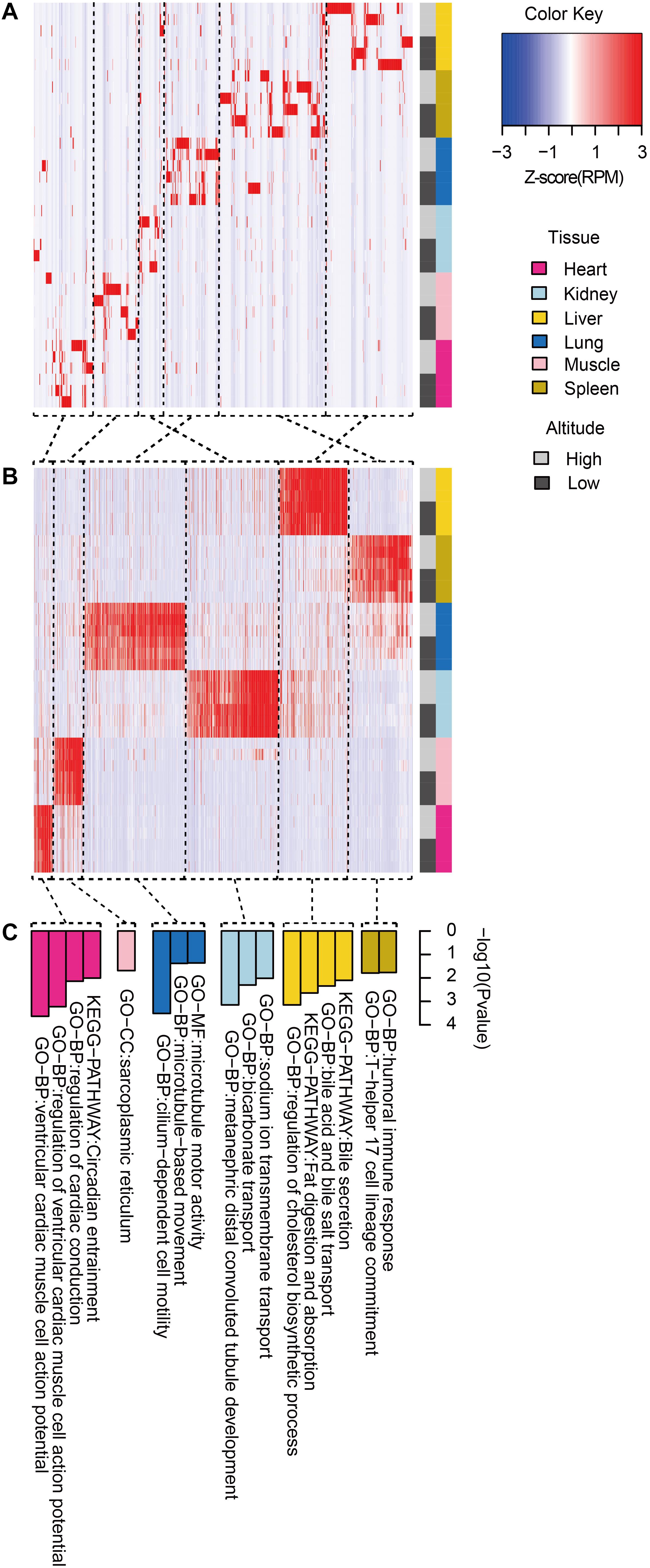
Figure 3. Functional pattern of tissue specificity for miRNAs and protein-coding genes. Hierarchical clustering of (A) miRNAs and (B) protein-coding genes. Average linkage hierarchical clustering was used, with the distance between transcripts measured by Pearson’s correlation between the vectors of the z-score standardized expression values. (C) Functional enrichment of tissue-specific genes targeted by tissue-specific miRNAs within corresponding tissues. Representative GO terms and KEGG pathways.
Comparison of miRNAs Between the High- and Low-Altitude Populations
To explore the transcriptional changes of miRNAs induced by acclimatization of goats to high-altitude stress, we performed comparisons between high- and low-altitude goat populations within each tissue. In total, we detected 138 differentially expressed mature miRNAs between high- and low-altitude populations (heart, 37; kidney, 20; liver, 40; lung, 26; muscle, 47; spleen, 37) (Table 2).
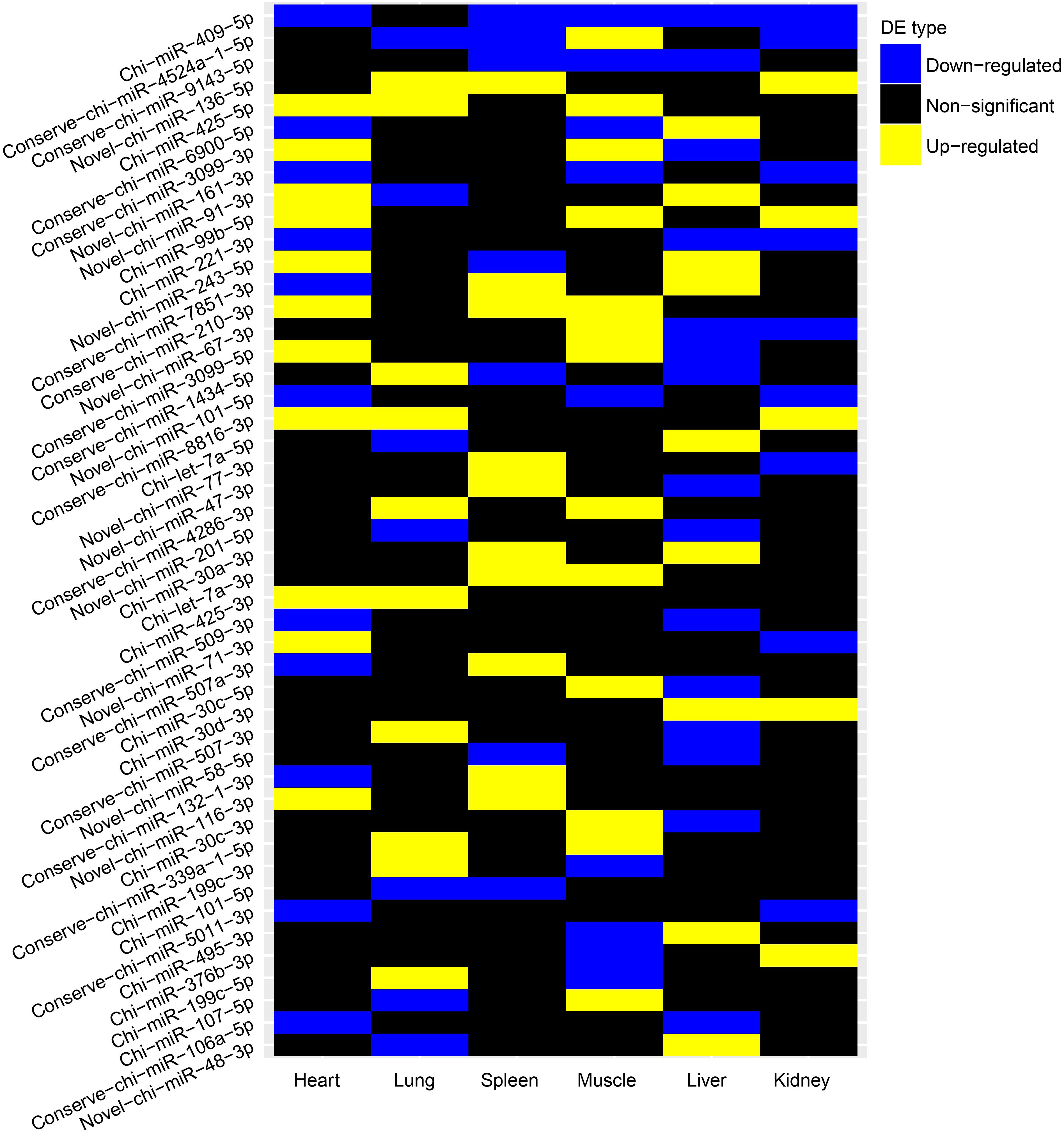
We further evaluated whether these differentially expressed (DE) miRNAs showed similar expression alteration patterns among the six tissues, because of the strong natural selection pressure on high-altitude goat populations. Tissue-specific analysis of DE miRNAs showed that miRNAs with expression changes between altitudes had a lower proportion of tissue-specific miRNAs than those that were not differentially expressed (Supplementary Figure S4). Evaluation of the overlap of expression alterations across tissues revealed that most of the 138 DE miRNAs (n = 91) underwent expression changes with no overlap among tissues, whereas the rest (n = 47) showed consistent up- or down-regulation among up to five tissues (Table 3). Interestingly, miRNAs differentially expressed in only one tissue were also significantly (Chi-squared test, P = 9.55 × 10–6) more tissue-specific (41/91) than those with expression alterations in more than one tissue (3/47) (Supplementary Figure S5). It is worth noting that those 41 tissue-specific non-overlapping DE miRNAs were generally enriched in tissue-related functions, such as calmodulin binding (nine genes, P = 7.20 × 10–3) in heart, regulation of ion transmembrane transport (five genes, P = 5.36 × 10–3) in kidney, and positive regulation of fibroblast proliferation (seven genes, P = 4.03 × 10–4) in muscle (Supplementary Table S2). Alternatively, the overlapping DE miRNAs functioned in more widespread processes in high-altitude acclimatization (Figure 4); for example, miR-409-5p was consistently down-regulated in five tissues (heart, kidney, liver, muscle, and spleen) of the high-altitude goat population, and its target genes were significantly enriched in hypoxia-related functions, such as positive regulation of angiogenesis and apoptosis.
DE miRNAs Involved in High-Altitude Acclimatization
To illustrate the potential functions of identified DE miRNAs on a larger scale, we performed target prediction on DE miRNAs and only maintained miRNA–mRNA pairs with negative correlation coefficients as high-confidence target pairs for further analysis. In particular, a considerable portion of high-confidence target genes were detected with known or potential roles in high-altitude-related responses, such as apoptosis, angiogenesis, DNA damage repair, erythropoiesis, and energy metabolism, which are highly related to high-altitude acclimatization according to previous studies (Supplementary Table S3).
Thereafter, as revealed by functional enrichment analysis of high-confidence target genes, the majority of target genes might have known or potential roles in hypoxia response among all six tissues (Figure 5). For example, target genes were significantly enriched in protein-ubiquitination related categories, including ubiquitin binding (75 genes, P = 5.5 × 10–6), ubiquitin-protein transferase activity (238 genes, P = 1.8 × 10–9), and regulation of proteasomal ubiquitin-dependent protein catabolic process (69 genes, P = 3.7 × 10–4), which serves as a critical component of hypoxia response. Additionally, target genes were also significantly enriched in categories related to DNA damage repair, such as mitotic G1 DNA damage checkpoint (43 genes, P = 2.7 × 10–2) and signal transduction involved in DNA damage checkpoint (40 genes, P = 2.0 × 10–2). Moreover, functional enrichment in positive regulation of apoptotic process (288 genes, P = 5.25 × 10–5) and angiogenesis (211 genes, P = 3.63 × 10–4) were also found in target genes shared by all six tissues, as were pathways such as HIF-1 (49 genes, P = 5.66 × 10–2), p53 (39 genes, P = 1.00 × 10–2), and insulin signaling pathways (86 genes, P = 1.09 × 10–6).
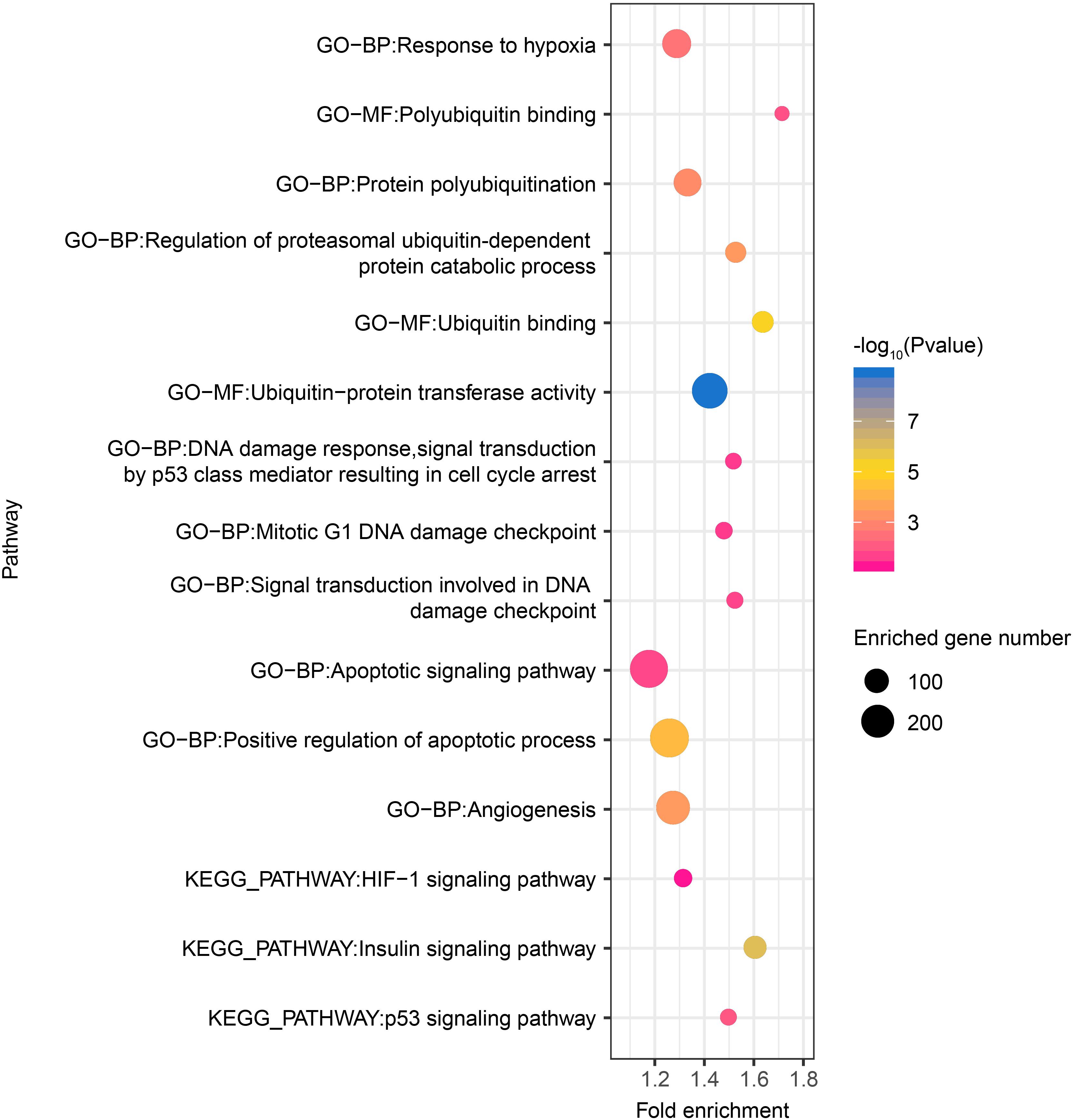
Figure 5. Hypoxia-related functional gene categories enriched by target genes of DE miRNAs among all six tissues between high- and low-altitude populations. The P value was calculated using the Benjamini-corrected modified Fisher’s exact test. BP, biological process; MF, molecular function; CC: cellular component.
By comparing mRNA expression alterations of these target genes between altitudes, we found more reliable evidence for acclimatization based on miRNA–mRNA interactions in hypoxia-related pathways. Our findings mainly focused on the interplay between DE miRNA and target genes, especially in the HIF-1 signaling pathway (Figure 6A). By collectively analyzing all six tissues, we found 102 pairs that consisted of 37 DE miRNAs with 31 target genes that were previously confirmed to be differentially expressed involved in both upstream and downstream of the HIF-1 signaling pathway (Supplementary Table S4). In heart, genes encoding both isoforms of phospholipase Cγ (PLCγ), PLCG1, and PLCG2 (Guoping et al., 2012), were found to be up-regulated by decrease of the DE miRNAs conserve-chi-miR-509-3p and conserve-chi-miR-3069-1-3p (PLCG1), and chi-miR-409-5p, conserve-chi-miR-3069-1-3p, and conserve-chi-miR-208a-3p (PLCG2). Genes that encode isoforms of the downstream Ca2t/calmodulin kinase II (CaMKII), CAMK2A and CAMK2D (Swulius and Waxham, 2008), were also up-regulated because of reduced expression of the DE miRNAs conserve-chi-miR-509-3p (CAMK2A) and conserve-chi-miR-509-3p, conserve-chi-miR-208a-3p, and conserve-chi-miR-3069-1-3p (CAMK2D) (Figure 6A). As expected, mRNA expression levels of HIF-1 target genes, including VEGFA, FLT1, Glut1, HK1, and TFRC, that were involved in angiogenesis, anaerobic metabolism, and iron metabolism were significantly up-regulated, and the transcription of FLT1 and TFRC were further activated by decrease of novel-chi-miR-232-3p, conserve-chi-miR-5011-3p, chi-miR-409-5p, and conserve-chi-miR-106a-5p (Figure 6A).
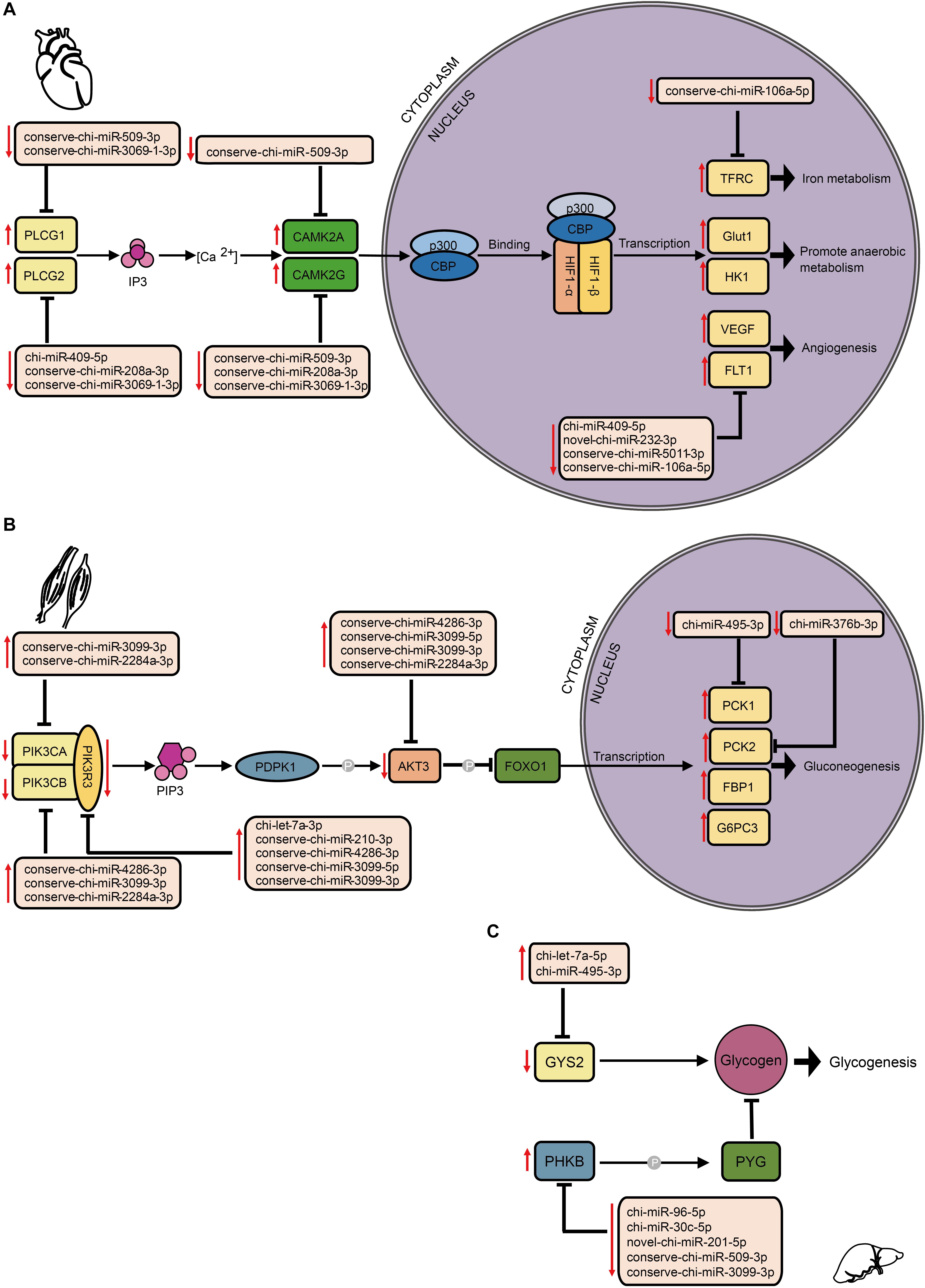
Figure 6. Schematic representation of hypoxia-related signaling pathways enriched by target genes of DE miRNAs in different tissues, including (A) the HIF-1 signaling pathway in heart, and the insulin signaling pathway in (B) muscle and (C) liver. The up and down arrows in red indicate up- and down-regulation, respectively, of DE miRNAs or genes. The clip art of tissues were created on our own.
In the insulin signaling pathway, we detected 103 pairs of DE miRNA–mRNA (Supplementary Table S4). In muscle, we found eight DE miRNAs for which their target genes correspondingly exhibited negative regulation. Upstream of the transcription factor FOXO1, genes encoding PI3K subunits and Akt were modulated down. Alternatively, the genes that encode PCK1 and PCK2 were up-regulated (Figure 6B). In liver, the expression levels of chi-miR-495-3p and chi-let-7a-5p significantly increased, and the target gene GYS2 had decreased expression. Moreover, the gene encoding phosphorylase kinase regulatory subunit beta (PHKB) was simultaneously up-regulated with the down-regulation of novel-chi-miR-201-5p, conserve-chi-miR-509-3p, chi-miR-96-5p, chi-miR-30c-5p, and conserve-chi-miR-3099-3p.
In the p53 signaling pathway, we found 69 pairs of DE miRNA–mRNA (Supplementary Table S4). Interestingly, PAI-1 and TSP1 were down-regulated, whereas their corresponding target miRNAs were up-regulated in liver, lung, and muscle. The down- and up-regulation of TSP1 and its target miRNA were also detected in heart and kidney. In addition, chi-miR-409-5p and chi-miR-450-3p in spleen were down-regulated, with increased expression of their target gene, RRM2B. Furthermore, up-regulation of several DE miRNAs was found along with down-regulation of their target genes, CDK6 in both kidney and muscle and CCND2 (kidney)/CCND3 (muscle) (Figure 7).
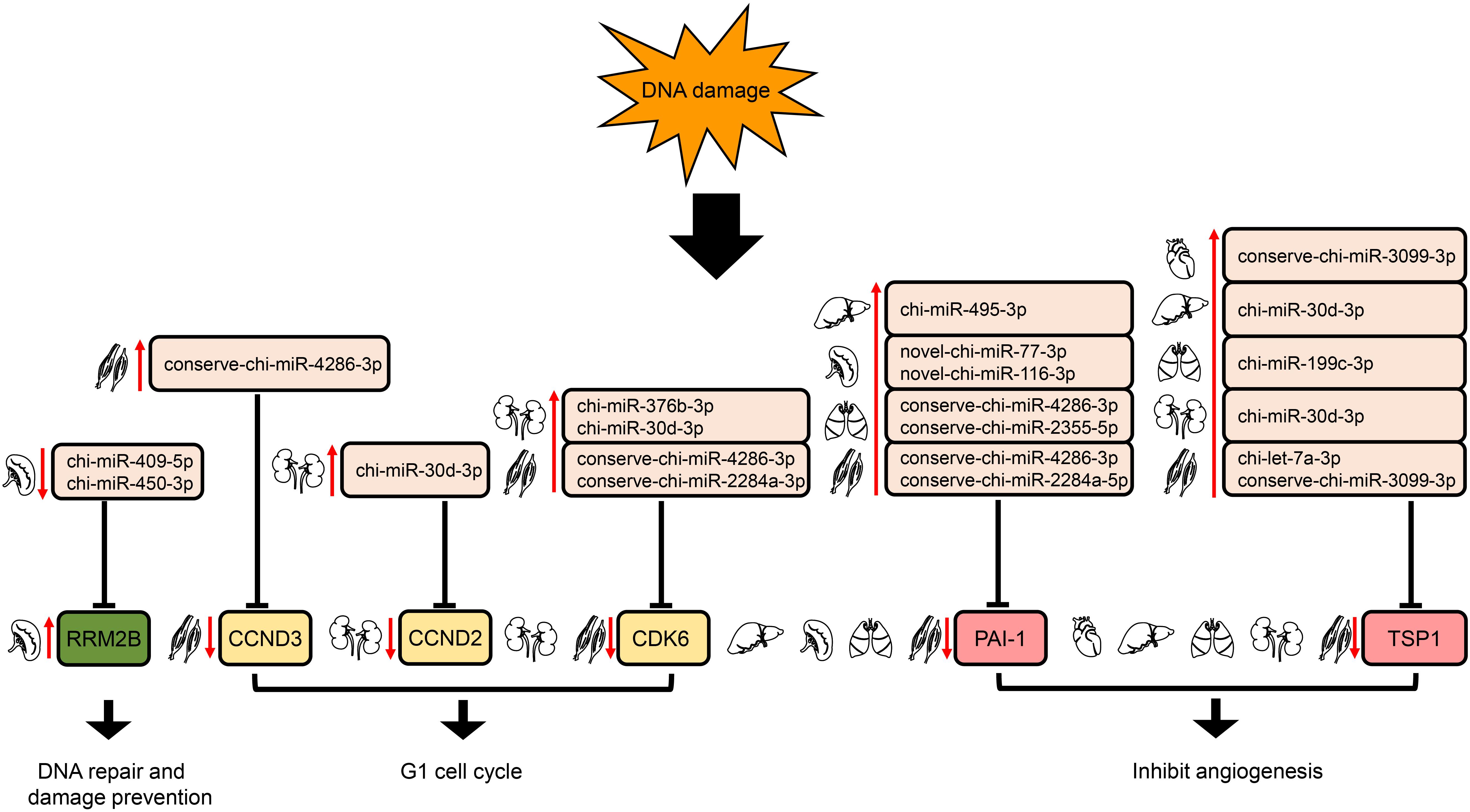
Figure 7. Regulation of genes targeted by DE miRNAs enriched in the downstream of p53 signaling pathway. The up- and down-arrows in red mean up- and down-regulation of DE miRNAs or genes, respectively. The clip art of tissues were created on our own.
As confirmed by qRT-PCR, the fold change of relative expression between low- and high-altitude were found significantly correlated with the fold change of normalized read counts in DE miRNAs (9/11, Supplementary Table S5), especially for those that are likely to play a crucial role in high-altitude acclimatization-related pathways. Next, we selected nine candidate mRNA–miRNA pairs and performed the dual luciferase reporter assays to validate the potential relationship of these pairs. As shown in Supplementary Figure S6, eight miRNAs significantly repressed luciferase activity in HeLa cells transfected with the 3’-UTR reporter, which demonstrated the robustness of mRNA–miRNA pairs in this study. Thus, these results provide robust evidence for high-altitude acclimatizations in domestic goats based on miRNA expression alterations.
MiR-106a-5p Had a Negative Regulation Effect on Angiogenesis
Based on the results of dual luciferase assays, we noticed that FLT-1 was the direct target gene of miR-106a-5p, and FLT-1, also named VEGFR1, were reported to be involved in angiogenesis (Hiratsuka et al., 2001; Shibuya, 2006). Thus, miR-106a-5p was selected as a representative for exploring the biological function of DE miRNAs involved in high-altitude acclimatization. Sequence alignment analysis showed that miR-106a-5p manifested high similarity especially in seed sequence among several representative species (Figure 8A). Meanwhile, the 3’-UTR of FLT-1 containing the miR-106a-5p binding site were also conserved among these species (Figure 8A). Given lack of commercial cell line in goat, we performed miRNA function analysis using the human umbilical venous endothelial cells (HUVECs), a widely used cell line for angiogenesis research. Effective inhibition of miR-106a-5p was confirmed using qRT-PCR (P < 0.01, Figure 8B). As the angiogenesis involves many key processes, including proliferation, migration and tube formation (Wang et al., 2011), we thus assessed them by CCK-8 detection, Edu staining, scratch assay and tube formation experiments. These results showed that miR-106a-5p downregulation promoted HUVECs proliferation (P < 0.01, Figures 8C,D), enhanced migration ability (P < 0.01, Figure 8E), and increased novel cellular junctions and tube-length (P < 0.01, Figure 8F), compared with NC. Meanwhile, the expression level of pro-angiogenesis genes including VEGF, FLT-1 and Notch-1 significantly increased, and that of anti-angiogenesis gene TSP1 decreased (P < 0.01, Figure 8G). These results indicated that miR-106a-5p had a negative regulation effect on angiogenesis.
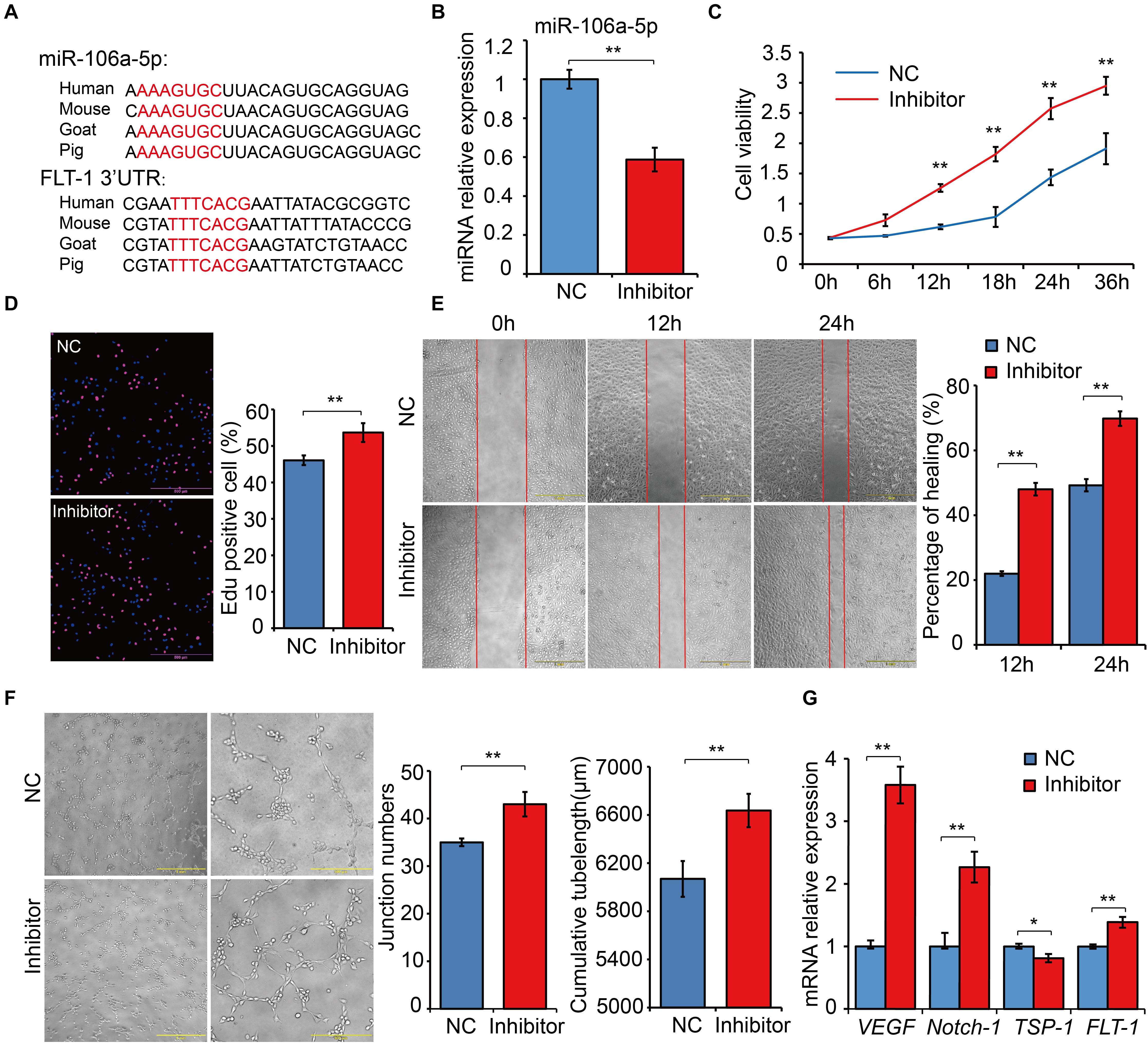
Figure 8. Functional experiments of miR-106a-5p regulation effect on angiogenesis. (A) Sequence alignment analysis of miR-106a-5p and the 3’-UTR of FLT-1. (B) miR-106a-5p inhibition efficiency analysis using qRT-PCR. miR-106a-5p inhibitor or NC was transfected into HUVECs, cell viability (C), proliferation (D), migration (E) and angiogenesis (F) were assessed by CCK-8 detection, Edu staining, scratch assay and tube formation experiments, respectively. (G) qRT-PCR quantification of pro-angiogenesis genes, including VEGF and Notch-1, and anti-angiogenesis gene (TSP-1) and the target gene of miR-106a-5p (FLT-1). At least three independent experiments were repeated three times. All values are presented as mean ± standard deviation (SD).
Discussion
To date, no studies have illustrated the role of goat miRNA in high-altitude acclimatization. Hence, to the best of our knowledge, this is the first report to compare goat miRNA profiles between altitudes and among multiple tissues.
As a result of tissue specificity analysis for miRNAs and protein-coding genes, miRNAs exhibited more tissue specificity than protein-coding genes in goats (Figure 2), which is consistent with the findings of a previous study on human (Ludwig et al., 2016). Further analysis for tissue-specific miRNA–mRNA target pairs showed clustering of both tissue-specific miRNA (Figure 3A) and mRNA (Figure 3B), which indicated the similar expression pattern of tissue-specific transcripts in each tissue. In particular, enrichment in tissue-related functional categories were found in clusters of tissue-specific genes targeted by tissue-specific miRNAs (Figure 3C). Therefore, the potential regulation of tissue-specific protein-coding genes by tissue-specific miRNAs may play a crucial role in tissue function maintenance.
As a result of miRNA differential expression, muscle exhibited the highest number of DE miRNAs among the six tissues. This highly dramatic transcriptional change in the muscle reflects that, among the six tissues, skeletal muscle cells were most sensitive to hypoxia and most closely associated with high-altitude acclimatization (Howald and Hoppeler, 2003; Murray, 2009; Levett et al., 2012).
Combining expression alterations and tissue specificity of miRNAs, we found that miRNAs with expression changes between altitudes were clearly less tissue-specific than those that were not differentially expressed (Supplementary Figure S4). Despite of this, most of the DE miRNAs underwent expression changes with no overlap among tissues, whereas the rest showed up- or down-regulation in multiple tissues (Table 4). Interestingly, miRNAs differentially expressed in only one tissue were also more tissue-specific than those with expression alterations in more than one tissue (Supplementary Figure S5). Further more, we found that tissue-specific non-overlapping DE miRNAs were generally enriched in tissue-related functions, whereas the overlapping DE miRNAs functioned in more widespread processes in high-altitude acclimatization (Figure 4). With consistent up/down-regulation in multiple tissues, the tumor suppressor miR-425 is able to repress the PI3K-Akt pathway by targeting IGF-1 (Liu et al., 2015). The key mediator of mTOR kinase, miR-99b, contributes to radiation-induced mTOR up-regulation, which plays a critical role in radio-resistance (Wei et al., 2013). miR-221 has the ability to block endothelial cell migration, proliferation, and angiogenesis in vitro by targeting c-Kit and indirectly regulating expression of eNOS (Angelika et al., 2008). miR-210 serves as the micromanager of the hypoxia pathway and regulates many hypoxia-related processes, such as angiogenesis, cell cycle regulation, and DNA damage repair (Huang et al., 2010). Taken together, the expression alterations and tissue-specificity of miRNAs among tissues suggested not only the potential tissue-specific mechanism to maintain normal tissue functions under hypoxia, but also the tissue-conserved miRNA acclimatization to high altitude.
Furthermore, literature search on high-confidence target genes of DE miRNAs indicated their roles in high-altitude-related biological processes (Supplementary Table S3). For example, the energy metabolism-related genes USP1, JAK2, MTPN, and PDPK1 were predicted targets of the DE miRNA conserve-chi-miR-375-3p in this study, which is consistent with previous research (Poy et al., 2004, 2009; El Ouaamari et al., 2008); similarly, IGF1R, INSR, and IRS2 were targeted by chi-let-7a-3p (Sun et al., 2009; Zhu et al., 2011). This finding is also well supported by functional enrichment of target genes (Figure 5). For instance, three classes of target genes were found significantly enriched in protein-ubiquitination related categories. Since HIF-1α protein degradation in hypoxia is ubiquitination-dependent, and E3 ubiquitin ligases may participate in this degradation pathway (Ivan et al., 2001; Jaakkola et al., 2001; Anna et al., 2004; Nakayama et al., 2009; Wang et al., 2016), these target genes are likely to participate in hypoxia response. Analogously, target genes were also found enriched in categories related to DNA damage repair. As it is common knowledge that high-altitude exposure results in decreased oxygen pressure and increased UV radiation, which lead to DNA damage (Dosek et al., 2007), we speculate that Tibetan goats might have evolved strong abilities to resist DNA damage caused by hypoxia and intense UV radiation during long-term acclimatization to the extreme environmental conditions in the Tibetan Plateau.
Through comparisons of miRNA–mRNA expression alterations between altitudes, we found considerable evidence for acclimatization based on miRNA–mRNA interactions in hypoxia-related pathways, including HIF-1, p53, and insulin signaling pathways. HIF-1 is a basic transcription factor that is expressed in all metazoan organisms and consists of HIF-1α and HIF-1β subunits, and it functions as a master regulator of oxygen homeostasis (Semenza, 2007). Under hypoxic conditions, HIF-1 regulates the transcription of numerous hypoxia-response genes involved in angiogenesis (Fraisl et al., 2009) and energy metabolism (Ke and Costa, 2006) by binding to hypoxia-response elements. Moreover, during high-altitude hypoxia, ROS production by the mitochondrial electron transport system can increase (Mohanraj et al., 1998), while increased levels of ROS have been shown to activate PLCγ (González-Pacheco et al., 2002), which generates inositol 1, 4, 5-trisphosphate (IP3). Activated IP3 receptors can lead to elevations in Ca2+ via mobilization of intracellular Ca2+ stores (Yuan et al., 2010), which activates CaMK (Ying-Jie et al., 2010) and thereby phosphorylates the HIF-1 co-activators p300 and CREB-binding protein (Hardingham and Arnold, 2001) to form an effective transcriptional complex that can regulate the transcription of target hypoxia-response genes (Azimi, 2018). In this study, the down-regulation of conserve-chi-miR-509-3p, conserve-chi-miR-3069-1-3p, chi-miR-409-5p and conserve-chi-miR-208a-3p in heart were possibly responsible for the expression elevation of genes encoding PLCγ and CaMK, which promote the transcriptional activity of HIF-1. Downstream of HIF-1 transcriptional complex, mRNA expression levels of several HIF-1 target genes that were involved in angiogenesis, anaerobic metabolism, and iron metabolism were found significantly up-regulated (Figure 6A). Notably, we confirmed that FLT-1 was a direct target gene of miR-106a-5p by dual luciferase reporter assay. FLT-1 is the receptor of VEGF (Hiratsuka et al., 2001; Shibuya, 2006) and plays important roles in endothelium angiogenesis (Wang et al., 2011). Functionally, miR-106a-5p downregulation could promote angiogenesis through promoting proliferation, enhancing migration ability, and increasing tube formation of endothelial cells by mitigating its inhibition effect on FLT-1, which further elucidated the potential mechanism of miRNA-mediated regulations in angiogenesis. Therefore, the expression alterations of miRNAs might play an important role in high-altitude acclimatization through the HIF-1 signaling pathway.
Insulin functions as the primary hormone involved in glucose homeostasis and the stimulation of glucose transport through the insulin signaling pathway (Saltiel and Kahn, 2001), which can be inhibited and switch to a state of insulin resistance under hypoxia (Claire et al., 2009). As previously reported, PI3K and Akt could inhibit the transcription factor FOXO1, and PCK1 and PCK2 are the key enzymes in gluconeogenesis. In muscle, the down-regulation of genes encoding PI3K subunits and Akt and the up-regulation of genes that encode PCK1 and PCK2 (Figure 6B) indicated enhanced glucose production via gluconeogenesis in response to hypoxia. GYS2 encodes the key isozyme of glycogenesis, whereas PHKB phosphorylates and activates glycogen phosphorylase (PYG) (Tung et al., 1984). Because PYG catalyzes the rate-limiting step in glycogenolysis and breaks up glycogen into glucose subunits (Newgard et al., 1989), the down-regulation of GYS2 and up-regulation of PHKB by corresponding DE miRNAs in liver (Figure 6C) suggested inhibited liver glycogenesis at high-altitude. Hence, DE miRNAs might participated in the regulation of glycometabolism via the insulin signaling pathway and help with energy expenditure under the severe environment at high altitude.
The p53 pathway is composed of hundreds of genes and their products that respond to a wide variety of stress signals, such as DNA damage and hypoxia (Levine et al., 2006). p53 activation triggers the transcription of numerous genes involved in cell cycle arrest, apoptosis, DNA repair, and anti-angiogenesis (Harris and Levine, 2005). In liver, lung, and muscle, the angiogenesis inhibitors (Rodriguez-Manzaneque et al., 2001; Skrzypczakjankun, 2001) PAI-1 and TSP1 were modulated down due to the up-regulation of their corresponding target miRNAs. Whereas in heart and kidney, TSP1 was down-regulated by the expression elevation of its target miRNA (Figure 7). Moreover, down-regulation of chi-miR-409-5p and chi-miR-450-3p in spleen likely increased expression of their target gene, RRM2B; this gene encodes the protein p53R2, which is necessary for DNA repair. In addition, up-regulation of several DE miRNAs might repress their target genes, CDK6 in both kidney and muscle and CCND2 (kidney)/CCND3 (muscle) (Figure 7); these genes encode the key proteins in G1/S checkpoint function (Pietenpol and Stewart, 2002). Therefore, DE miRNAs involved in the p53 signaling pathway might play a significant role in responses to high-altitude environmental stress.
Despite the evidences we found in regulatory mechanisms of high-altitude acclimatization in domestic goats, we must admit the limitation that breed differences were not fully excluded in this comparative miRNA transcriptome analysis. It is of great interest to conduct the experimental design of moving the indigenous goat to high altitude for years and to better illustrate the mechanisms of high-altitude acclimatization on the same breed level.
Overall, based on our attractive goat transcriptomic resource, future work could be done on both in silico and experimental sides to improve our knowledge on high-altitude acclimatizations in goat populations: to further illustrate relationships among high-altitude acclimatization-related signaling pathways (Figures 6, 7) by conducting network analysis on involved miRNA–mRNA interactions, and to experimentally validate functions of highlighted miRNAs whose target genes has already been validated by our luciferase reporter assays (Supplementary Figure S6). Accompanied by the continuous development of goat genome sequence resources, fundamental questions of evolutionary adaptation in goats could be addressed in a more comprehensive way, by integrating phenotypic, population genomic and transcriptomic data.
Conclusion
This study presents a comprehensive and systematic survey of miRNA transcriptome in goat populations at high- and low-altitude, shedding light on the complicated miRNA expression patterns and associated regulatory mechanisms of high-altitude acclimatization in domestic goats. We expanded annotation of goat miRNAs to more than three times as many as the miRBase annotation using small RNA-seq data. Combination analysis of expression alterations and tissue specificity of miRNAs suggested potential coexisting tissue-specific and -conserved mechanisms for hypoxia acclimatization. Integrated miRNA-mRNA expression profiles and functional enrichment further provided evidence of miRNA involvements in post-transcriptional regulation through HIF-1, insulin, and p53 signaling pathways, which might promote anaerobic metabolism, angiogenesis, gluconeogenesis, DNA damage repair and inhibit glycogenesis. In addition, we experimentally confirmed miR-106a-5p to have a negative regulation effect on angiogenesis by directly targeting FLT-1. Taken together, these findings support the viewpoint that miRNAs could play significant roles in high-altitude acclimatization of domestic goats. Our study may not only accelerate research into goats as natural models for studying high-altitude acclimatization, but also provide valuable theoretical underpinnings for further utilization of genetic resources in plateau regions.
Materials and Methods
Animals and Samples Collection
To control breed differences which might contribute to overall observed differences, we selected goat breeds under the following rules: (1) Located in geographically close regions within 300 km straight-line distance and (2) Without significant geographic isolation. Following these rules, we selected the indigenous goat and Tibetan goat residing at distinct altitudes in southwest China (600 and 3000 m) for comparative analysis, between which the straight-line distance was 299 km. Three adult females (∼3 years old) from each of the indigenous goat and Tibetan goat populations residing at distinct altitudes in southwest China (600 and 3000 m) were used in this study (Table 4). For the home-reared indigenous goats, we selected the individuals unrelated within three generations according to the pedigree; for the stocking Tibetan goats, we randomly selected three female individuals. All the animals were provided by local farm. To control the diet differences between rearing modes of home rearing and stocking/nomadism, we fed the home-rearing Indigenous goat with natural pasture grass instead of artificially added compound feeds. Animals were fed with free access to food and water, and killed humanely to ameliorate suffering by putting them under deep anesthesia with intravenous sodium pentobarbital (30 mg/kg body weight) before slaughter. Six typical hypoxia-sensitive tissues (heart, kidney, liver, lung, longissimus muscle, and spleen) were rapidly excised from the carcass, immediately frozen in liquid nitrogen, and then stored at −80°C until RNA isolation.
Research procedures involving animals were performed according to the Regulations for the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China, revised in June 2004) and approved by the Institutional Animal Care and Use Committee in College of Animal Science and Technology, Sichuan Agricultural University, Sichuan, China under permit No. DKY-S20163658.
RNA Isolation, Small RNA Library Preparation and Sequencing
Total RNA was extracted using Trizol reagent (Ambion, United States) according to the manufacturer’s protocols. Small RNA libraries were constructed using the Illumina TruSeq Small RNA Sample Prep kit. Libraries were assessed using the Agilent 2200 TapeStation and sequenced on the Illumina HiSeq 2500 platforms. Incipient bioinformatics analysis (base calling) was performed with CASAVA 1.8 (Illumina) to generate raw reads (in FASTQ form).
Data Downloading and Processing for mRNA Transcriptome
To elucidate the effects of miRNA on downstream expression, we downloaded 36 additional transcriptome data sets of the exactly corresponding goat samples from our previous study (Tang et al., 2015) stored in the Gene Expression Omnibus (GEO) under accession code GSE66242. After a strict quality control, high-quality reads obtained were mapped to the representative goat genome (assembly ARS1) using hisat version 2.0.5 (Kim et al., 2015). Stringtie version 1.3.3 was used to quantify gene expression and obtain FPKM (denoted as fragments per kilobase of exon per million fragments mapped) expression values (Mihaela et al., 2015). We further used Cuffdiff 2 (Cole et al., 2013) to detect differentially expressed genes (DEGs) between population pairs from distinct altitudes in the six tissues. We defined genes with |log2 (fold change)| ≥ 1, P value <0.05 and adjusted P value <0.1 as DEGs. Differentially expressed genes with a log2 (fold change) < 0 were defined as “up-regulated” and those with a log2 (fold change) > 0 were defined as “down-regulated” at high altitude.
Read Mapping and miRNA Identification
Raw reads was subjected to a series of stringent filters (such as removing low quality-reads, repeated sequences and adaptor sequences). Filtered high-quality sequences were then mapped to goat reference genome (assembly ARS1) with stringent criteria (0 mismatch in the whole length) using Bowtie (Langmead, 2010). The number of mappable reads were similar between high and low altitudes (Supplementary Figure S2); therefore, the miRNA libraries had unbiased sizes for further analysis. Next, mappable reads were submitted to miRDeep version 2.0.0.7 (Friedlander et al., 2012) to detect miRNAs with default parameters, while mature miRNA sequences of goat and all other annotated mammalian species in miRbase release 21 (Kozomara and Griffiths-Jones, 2014) were selected as reference. miRNAs with read count no less than 3 in at least one sample were remained for further analysis. Read counts were normalized by the total count of mappable reads of each sample, also known as RPM, for unbiased comparisons among samples. Normalized expressions were further standardized by computing standard scores (also known as z-scores) for cluster and PCA (Wichern and Johnson, 1982; Everitt and Hothorn, 2011).
Qualitative and Quantitative Measure for Tissue Specificity
To evaluate the variability of expression patterns, we brought in two measures, TSI (Ludwig et al., 2016) and TSS (Cabili et al., 2011), which respectively indicate whether a transcript is tissue-specific and to which tissue this transcript is most tissue-specific.
The TSI is a quantitative measure for the specificity of a transcript in different tissues. The values range from 0 to 1. The higher a TSI, the more tissue-specific a transcript is. In this study, we defined a transcript with a TSI < 0.15 as “housekeeping” and >0.85 as “tissue-specific.”
The TSS is an entropy-based measure that quantifies the similarity between a transcript’s expression pattern and another predefined pattern that represent an extreme case in which a transcript is expressed in only one tissue. For each transcript, a TSS was calculated with respect to each tissue and the tissue with the maximal TSS was considered specific (e.g., heart-specific) if the TSI of this transcript is larger than 0.85.
Identification of DE miRNAs
To compare the expression levels of miRNA transcriptome between low-altitude and Tibetan goat, we identified DE miRNAs using edgeR package (Robinson et al., 2010; Mccarthy et al., 2012). Read counts were loaded into edgeR and normalized using the built-in trimmed mean of M-values (TMM) algorithm. miRNAs with a |log2 (fold change)| ≥ 1.5 and a Benjamini Hochberg FDR < 0.05 between altitudes were defined as DE miRNAs. DE miRNAs with a log2 (fold change) < 0 were defined as “up-regulated” and those with a log2 (fold change) > 0 were defined as “down-regulated” at high altitude.
Target Prediction and Functional Annotation of miRNAs
We applied TargetScan version 7.0 to DE miRNAs and 3’UTR sequences extracted from goat genome for target predications (Agarwal et al., 2015). Thereafter, we calculated Spearman’s correlations for expression levels of every miRNA-mRNA candidate target pair and maintained pairs with negative correlation coefficients as high-confidence target genes. The enrichment of GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway by target genes were performed using DAVID web-accessible program (Da et al., 2009) (Benjamini adjusted P value ≤0.05).
Quantitative Real-Time PCR Validation
To validate gene expression, quantitative real-time PCR (qRT-PCR) was conducted on 11 miRNAs in tissue samples from high- and low-altitude goat individuals, 1 miRNA in HUVECs, and 4 mRNAs in HUVECs. miRNAs and mRNAs were reverse transcribed using a Mir-XTM miRNA First Strand Synthesis Kit (Takara, Dalian, China) and PrimeScript RT reagent Kit with gDNA Eraser (Takara, Dalian, China), respectively. Quantitative real-time PCR was performed using a SYBR Premix Ex Taq kit (Takara, Dalian, China) on the CFX96TM Real-Time PCR Detection System (Bio-Rad, CA, United States). U6 and GAPDH were simultaneously used as endogenous control genes for miRNAs and mRNAs, respectively. And primer sequences for qRT-PCR are shown in Supplementary Table S6. A negative control (NC) was introduced into all measurements (no cDNA template), and each RNA sample was analyzed in triplicate. We computed relative expression levels of objective miRNAs with the 2–ΔΔCt method. To insure the accuracy of the 2–ΔΔCt results, we carried out the optimization of annealing temperature by thermal gradient and evaluation of amplification efficiency in CFX96TM Real-Time PCR Detection System (Bio-Rad, CA, United States), and all wells used in the subsequent analysis were met 0.95 < AE < 1.05, including the target genes and endogenous controls.
To confirm the differential expressions of selected DE miRNAs, we first calculated the fold change between low- and high-altitude goat populations using both relative expression and TMM-normalized read counts. Next, we calculated the Pearson’s correlations between fold change of relative expression and TMM-normalized read counts. miRNAs with a Pearson’s correlation <0.05 were confirmed as differential expressed.
Luciferase Reporter Assay
Based on identified high-confidence target pairs, we selected 9 DE miRNA–mRNA pairs, including miR-409-5p/PLCG2, miR-409-5p/CDK6, miR-409-5p/FLT1, miR-210-3p/PIK3R3, miR-106a-5p/FLT1, miR-509-3p/CAMK2G, miR-2355-5p/SERPINE1, miR-208a-3p/CAMK2G, and miR-30c-5p/PHKB for target validations. Luciferase activity assays were performed to validate the potential relationship between miRNAs and their target genes. In detail, 3’-UTR sequences containing the miRNA binding site of nine candidate target genes were synthesized and then inserted into the multiple cloning site of pmirGLO plasmid. The recombinant pmirGLO vector and miRNA mimic or NC was cotransfected into HeLa cells at 60% confluency using Lipofectamine 3000 (Invitrogen, Grand Island, NY, United States), according to the manufacturer’s instructions. After transfection for 48 h, dual luciferase activity was tested by Luciferase Dual Assay Kit (Promega, Madison, WI, United States). Luciferase activity was expressed as an adjusted value (firefly normalized to renilla).
Cell Culture and miRNA Transfection
Human umbilical venous endothelial cells were obtained from the cell bank of the Chinese Academy of Sciences and routinely maintained in DMEM (Hyclone, Logan, UT, United States) supplemented with 10% FBS (GIBCO, Grand Island, NY, United States) at 37°C, 5% CO2. According to experimental requirements, the specific inhibitor of miR-106a-5p and NC were purchased from RiboBio (Guangzhou, China). Transfection of miRNAs into HUVECs at 70% confluency was performed using Hiperfect (QIAGEN, Germany) in accordance with the manufacturer’s protocol. After 6 h in transfection, all groups were replaced with new medium for subsequent experimentation.
Scratch and Tube Formation Assay
Human umbilical venous endothelial cells were cultured to near confluence in 12-well plates. Cell monolayer were straight scratched with a 10 μL pipette tip. PBS gently washed three times to remove the non-adherent cells and cell debris in supernatants. Next, miR-106a-5p and NC were transfected into HUVECs. Photos were taken at the same fields of view at 0, 12, and 24 h after scratching using an Olympus IX53 microscope (Olympus, Tokyo, Japan). For tube formation analysis, the transfected HUVECs were collected and added to the matrigel pre-polymerized well in the 24-well plate. The de novo formed capillary-like structures were imaged and quantified using ImageJ software (Bethesda, MA, United States).
Cell Viability and Proliferation
Cell viability and proliferation were assessed using a Cell Counting kit 8 (Beyotime Biotechnology, Guangzhou, China) and Edu staining (RiboBio, Guangzhou, China) analysis according to manufacturer’s protocol, respectively. For CCK8 detection, 10 μL CCK8 reagent was added to the culture medium of the transfected HUVECs. After incubation for 4 h, OD450 were measured using a microplate reader (Thermo Fisher Scientific, Madrid, Spain). For Edu analysis, the transfected HUVECs were treated with 10 μM Edu for 24 h and incubated for 14 h. Edu staining was performed according to the manufacturer’s protocol, cell nuclei stained with DAPI. Images were captured using an Olympus IX53 microscope (Olympus, Tokyo, Japan). The ratio of Edu positive cells were calculated from three independent experiments. The data were expressed as mean ± SD.
Data Availability Statement
The datasets generated for this study can be found in the GEO (GSE125665 and GSE66242).
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee in College of Animal Science and Technology, Sichuan Agricultural University.
Author Contributions
SF, QT, and ML formulated and evolved the overarching research goals and aims. SF, JM, and QT developed and designed the methodology. SF completed all of the data analysis and was a major contributor in writing the manuscript. JM and KL performed the qRT-PCR validation. JZ, WQ, YL, LJ, XW, AJ, LL, and WX provided reagents, materials, laboratory samples, instrumentation and contributed to acquisition of animal information. SF, JM, and KL wrote the original manuscript. JM and ML reviewed and edited the manuscript. QT and ML took oversight and leadership responsibility for the research activity planning and execution. ML acquired the financial support for the project leading to this publication. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31872335, 31601918, 31530073, and 31772576), the National Key R&D Program of China (2018YFD0500403 and 2018YFD0501204), the Sichuan Province & Chinese Academy of Sciences of Science & Technology Cooperation Project (2017JZ0025), and the Key Project of Sichuan Education Department (16ZA0025). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00809/full#supplementary-material
FIGURE S1 | Length distribution of sequenced high quality reads for each sample. Each curve represents a sample.
FIGURE S2 | Number of mappable reads of samples from high- and low-altitude goat populations. Each dot represents a sample. Red and green boxes indicate high- and low-altitude samples, respectively.
FIGURE S3 | Genomic sources of miRNA precursor sequences. Each proportion of this pie chart and the corresponding color represents a type of genomic elements. ‘Ambiguous’ refers to miRNA precursor sequences that locate in both introns and exons due to alternative splicing of protein-coding genes. The percentage beneath the black line represented the expected proportion based on the fraction of the genome that corresponds to the considered genomic element, and the P value was calculated by permutation test.
FIGURE S4 | Distribution curve for the frequency of TSI of (A) DE miRNAs and (B) non-DE miRNAs.
FIGURE S5 | Number of DE miRNAs with tissue-specificity and overlap in tissues. The P value was calculated using the Pearson’s Chi-squared test with Yates’ continuity correction.
FIGURE S6 | Dual luciferase reporter assay performed on nine candidate miRNA-mRNA pairs to validate the potential target relationship.
TABLE S1 | RNA quality of all samples. The RNA integrity number (RIN) was used for determining RNA quality.
TABLE S2 | Functional enrichment of genes targeted by tissue-specific miRNAs which differentially expressed with no overlap among tissues.
TABLE S3 | High-confidence target genes participated in high-altitude-related response according to the literature.
TABLE S4 | Expression alteration of miRNAs and target genes involved in pathways related to high-altitude acclimatization.
TABLE S5 | qRT-PCR validations of DE miRNAs involved in pathways related to high-altitude acclimatization. The Pearson’s correlations were calculated between fold change (relative expression) and fold change (TMM-normalized read counts). The fold changes were calculated between low- and high-altitude goat populations within each tissue.
TABLE S6 | Primer sequences for qRT-PCR.
Abbreviations
DE, differentially expressed; DEGs, differentially expressed genes; FPKM, fragments per kilobase of exon per million fragments mapped; GEO, Gene Expression Omnibus; GO, Gene Ontology; HUVECs, human umbilical venous endothelial cells; KEGG, Kyoto Encyclopedia of Genes and Genomes; NC, negative control; PCA, principal components analysis; qRT-PCR, quantitative real-time PCR; RIN, RNA integrity number; RPM, reads per million; TMM, trimmed mean of M-values; TSI, tissue specificity index; TSS, tissue specificity score; UV, ultraviolet radiation.
References
Agarwal, V., Bell, G. W., Nam, J. W., and Bartel, D. P. (2015). Predicting effective microRNA target sites in mammalian mRNAs. eLife 4:e05005. doi: 10.7554/eLife.05005
Ai, H., Yang, B., Li, J., Xie, X., Chen, H., and Ren, J. (2014). Population history and genomic signatures for high-altitude adaptation in Tibetan pigs. BMC Genomics 15:834. doi: 10.1186/1471-2164-15-834
Al-Fageeh, M. B., and Smales, C. M. (2006). Control and regulation of the cellular responses to cold shock: the responses in yeast and mammalian systems. Biochem. J. 397, 247–259. doi: 10.1042/BJ20060166
Anamaria, N., and Henrik, K. (2014). Evolutionary dynamics of coding and non-coding transcriptomes. Nat. Rev. Genet. 15, 734–748. doi: 10.1038/nrg3802
Angelika, K., Carmen, U., and Stefanie, D. (2008). Targeting microRNA expression to regulate angiogenesis. Trends Pharmacol. Sci. 29, 12–15. doi: 10.1016/j.tips.2007.10.014
Anna, D. C., Roberta, D. M., Fabio, M., Giulio, P., Capogrossi, M. C., and Antonia, G. (2004). Hypoxia inhibits myogenic differentiation through accelerated MyoD degradation. J. Biol. Chem. 279, 16332–16338. doi: 10.1074/jbc.m313931200
Azimi, I. (2018). The interplay between HIF-1 and calcium signalling in cancer. Int. J. Biochem. Cell Biol. 97, 73–77. doi: 10.1016/j.biocel.2018.02.001
Baroukh, N., Ravier, M. A., Loder, M. K., Hill, E. V., Bounacer, A., Scharfmann, R., et al. (2007). MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. J. Biol. Chem. 282, 19575–19588. doi: 10.1074/jbc.M611841200
Barroga, C. F., Pham, H., and Kaushansky, K. (2008). Thrombopoietin regulates c-Myb expression by modulating micro RNA 150 expression. Exp. Hematol. 36, 1585–1592. doi: 10.1016/j.exphem.2008.07.001
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Berezikov, E. (2011). Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 12, 846–860. doi: 10.1038/nrg3079
Bigham, A. W., Wilson, M. J., Julian, C. G., Kiyamu, M., Vargas, E., Leon-Velarde, F., et al. (2013). Andean and Tibetan patterns of adaptation to high altitude. Am. J. Hum. Biol. 25, 190–197. doi: 10.1002/ajhb.22358
Bonci, D., Coppola, V., Musumeci, M., Addario, A., D’Urso, L., Collura, D., et al. (2008). The Mir-15a/Mir-16-1 cluster controls prostate cancer progression by targeting multiple oncogenic activities. Nat. Med. 14, 1271–1277. doi: 10.1038/nm.1880
Breen, E., Tang, K., Olfert, M., Knapp, A., and Wagner, P. (2008). Skeletal muscle capillarity during hypoxia: VEGF and its activation. High Alt. Med. Biol. 9, 158–166. doi: 10.1089/ham.2008.1010
Brookfield, H., and Allen, B. (1989). High-altitude occupation and environment. Mot. Res. Dev. 9, 201–209.
Brutsaert, T. D., Soria, R., Caceres, E., Spielvogel, H., and Haas, J. D. (1999). Effect of developmental and ancestral high altitude exposure on chest morphology and pulmonary function in Andean and European/North American natives. Am. J. Hum. Biol. 110, 383–395. doi: 10.1002/(sici)1520-6300(1999)11:3<383::aid-ajhb9>3.0.co;2-x
Cabili, M. N., Cole, T., Loyal, G., Magdalena, K., Barbara, T. V., Aviv, R., et al. (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927. doi: 10.1101/gad.17446611
Calin, G. A., Ferracin, M., Cimmino, A., Di, L. G., Shimizu, M., Wojcik, S. E., et al. (2005). A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Eng. J. Med. 353, 1793–1801.
Camps, C., Buffa, F. M., Colella, S., Moore, J., Sotiriou, C., Sheldon, H., et al. (2008). hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin. Cancer Res. 93, 325–334.
Chan, S. Y., Ying-Yi, Z., Craig, H., Mahoney, C. E., Zweier, J. L., and Joseph, L. (2009). MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metabol. 10, 273–284. doi: 10.1016/j.cmet.2009.08.015
Chen, C. H., Liu, Y. F., Lee, S. D., Huang, C. Y., Lee, W. C., Tsai, Y. L., et al. (2009). Altitude hypoxia increases glucose uptake in human heart. High Alt. Med. Biol. 10, 83–86. doi: 10.1089/ham.2008.1064
Chen, Y., and Gorski, D. H. (2008). Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood 111, 1217–1226. doi: 10.1182/blood-2007-07-104133
Cheviron, Z. A., and Brumfield, R. T. (2012). Genomic insights into adaptation to high-altitude environments. Heredity 108, 354–361. doi: 10.1038/hdy.2011.85
Claire, R., Pascal, P., Thierry, G., Rosanna, N. L., Issam, B. S., Mireille, C., et al. (2009). Hypoxia decreases insulin signaling pathways in adipocytes. Diabetes 58, 95–103. doi: 10.2337/db08-0457
Cohen, E. E., Zhu, H., Lingen, M. W., Martin, L. E., Kuo, W. L., Choi, E. A., et al. (2009). A feed-forward loop involving protein kinase Calpha and microRNAs regulates tumor cell cycle. Cancer Res. 69, 65–74. doi: 10.1158/0008-5472.can-08-0377
Cole, T., Hendrickson, D. G., Martin, S., Loyal, G., Rinn, J. L., and Lior, P. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53. doi: 10.1038/nbt.2450
Colleoni, F., Padmanabhan, N., Yung, H. W., Watson, E. D., Cetin, I., Tissot van Patot, M. C., et al. (2013). Suppression of mitochondrial electron transport chain function in the hypoxic human placenta: a role for miRNA-210 and protein synthesis inhibition. PLoS One 8:e55194. doi: 10.1371/journal.pone.0055194
Crosby, M. E., Kulshreshtha, R., Ivan, M., and Glazer, P. M. (2009). MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 69, 1221–1229. doi: 10.1158/0008-5472.CAN-08-2516
Cuesta, R., Martínezsánchez, A., and Gebauer, F. (2009). miR-181a regulates cap-dependent translation of p27kip1 mRNA in myeloid cells. Mol. Cell. Biol. 29, 2841–2851. doi: 10.1128/mcb.01971-08
Da, W. H., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Dan, Z., Chu, Y., Hu, W., and Ge, R. (2010). Expression of high altitude hypoxia on heme oxygenase-1 in rats kidney. J. Qinghai Med. Coll.
Davalos, A., Goedeke, L., Smibert, P., Ramirez, C. M., Warrier, N. P., Andreo, U., et al. (2011). miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc. Natl. Acad. Sci. U.S.A. 108, 9232–9237. doi: 10.1073/pnas.1102281108
Dejean, E., Renalier, M. H., Foisseau, M., Agirre, X., Joseph, N., Paiva, G. R. D., et al. (2011). Hypoxia-microRNA-16 downregulation induces VEGF expression in anaplastic lymphoma kinase (ALK)-positive anaplastic large-cell lymphomas. Leukemia 25, 1882–1890. doi: 10.1038/leu.2011.168
Dosek, A., Ohno, H., Acs, Z., Taylor, A. W., and Radak, Z. (2007). High altitude and oxidative stress. Respir. Physiol. Neurobiol. 158, 128–131.
Eichner, L. J., Perry, M. C., Dufour, C. R., Bertos, N., Park, M., St-Pierre, J., et al. (2010). miR-378(∗) mediates metabolic shift in breast cancer cells via the PGC-1beta/ERRgamma transcriptional pathway. Cell Metab. 12, 352–361. doi: 10.1016/j.cmet.2010.09.002
el Azzouzi, H., Leptidis, S., Dirkx, E., Hoeks, J., van Bree, B., Brand, K., et al. (2013). The hypoxia-inducible microRNA cluster miR-199a~214 targets myocardial PPARdelta and impairs mitochondrial fatty acid oxidation. Cell Metab. 18, 341–354. doi: 10.1016/j.cmet.2013.08.009
El Ouaamari, A., Baroukh, N., Martens, G. A., Lebrun, P., Pipeleers, D., and van Obberghen, E. (2008). miR-375 targets 3’-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes 57, 2708–2717. doi: 10.2337/db07-1614
Esau, C., Davis, S., Murray, S. F., Yu, X. X., Pandey, S. K., Pear, M., et al. (2006). miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3, 87–98. doi: 10.1016/j.cmet.2006.01.005
Esau, C., Kang, X., Peralta, E., Hanson, E., Marcusson, E. G., Ravichandran, L. V., et al. (2004). MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 279, 52361–52365. doi: 10.1074/jbc.C400438200
Esguerra, J. L., Bolmeson, C., Cilio, C. M., and Eliasson, L. (2011). Differential glucose-regulation of microRNAs in pancreatic islets of non-obese type 2 diabetes model Goto-Kakizaki rat. PLoS One 6:e18613. doi: 10.1371/journal.pone.0018613
Everitt, B., and Hothorn, T. (2011). An Introduction to Applied Multivariate Analysis with R. Berlin: Springer Science & Business Media.
Felli, N., Pedini, F., Romania, P., Biffoni, M., Morsilli, O., Castelli, G., et al. (2009). MicroRNA 223-dependent expression of LMO2 regulates normal erythropoiesis. Haematologica 94, 479–486. doi: 10.3324/haematol.2008.002345
Fish, J. E., Santoro, M. M., Morton, S. U., Yu, S., Yeh, R. F., Wythe, J. D., et al. (2008). miR-126 regulates angiogenic signaling and vascular integrity. Dev. Cell. 15, 272–284. doi: 10.1016/j.devcel.2008.07.008
Fornari, F., Gramantieri, L. M., Veronese, A., Sabbioni, S., Calin, G., Grazi, G., et al. (2008). MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 27, 5651–5661. doi: 10.1038/onc.2008.178
Fraisl, P., Mazzone, M., Schmidt, T., and Carmeliet, P. (2009). Regulation of angiogenesis by oxygen and metabolism. Dev. Cell. 16, 167–179. doi: 10.1016/j.devcel.2009.01.003
Friedlander, M. R., Mackowiak, S. D., Li, N., Chen, W., and Rajewsky, N. (2012). miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40, 37–52. doi: 10.1093/nar/gkr688
García-Hjarles, M. A. (1989). Sperm count and seminal biochemistry of high altitude inhabitants and patients with chronic altitude sickness. Arch. Biol. Med. Exp. 22, 61–67.
Gerin, I., Bommer, G. T., McCoin, C. S., Sousa, K. M., Krishnan, V., and MacDougald, O. A. (2010). Roles for miRNA-378/378∗ in adipocyte gene expression and lipogenesis. Am. J. Physiol. Endocrinol. Metab. 299, E198–E206. doi: 10.1152/ajpendo.00179.2010
González-Pacheco, F. R., Carlos, C., Maria Angeles, C., Deudero, J. J. P., Javier, A., Susana, Y., et al. (2002). Mechanism of vascular smooth muscle cells activation by hydrogen peroxide: role of phospholipase C gamma. Nephrol. Dial. Transpl. 17, 392–398. doi: 10.1093/ndt/17.3.392
Gramantieri, L., Ferracin, M., Fornari, F., Veronese, A., Sabbioni, S., Liu, C. G., et al. (2007). Cyclin G1 Is a Target of miR-122a, a MicroRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 67, 6092–6099. doi: 10.1158/0008-5472.can-06-4607
Guoping, F., Yuhong, C., James, S., Demin, W., and Renren, W. (2012). Phospholipase Cγ2 plays a role in TCR signal transduction and T cell selection. J. Immunol. 189, 2326–2332. doi: 10.4049/jimmunol.1103458
Hardingham, G. E., and Arnold, F. H. (2001). Nuclear calcium signaling controls CREB-mediated gene expression triggered by synaptic activity. Nat. Neurosci. 4, 261–267. doi: 10.1038/85109
Harris, S. L., and Levine, A. J. (2005). The p53 pathway: positive and negative feedback loops. Oncogene 24, 2899–908.
He, A., Zhu, L., Gupta, N., Chang, Y., and Fang, F. (2007). Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol. Endocrinol. 21, 2785–2794. doi: 10.1210/me.2007-0167
Hiratsuka, S., Maru, Y., Okada, A., Seiki, M., Noda, T., and Shibuya, M. (2001). Involvement of Flt-1 tyrosine kinase (vascular endothelial growth factor receptor-1) in pathological angiogenesis. Cancer Res. 61, 1207–1213.
Hitoo, N., Koh, O., Yoshitaka, I., Takahiro, H., Kazuya, N., Genzou, T., et al. (2010). MicroRNA-15b modulates cellular ATP levels and degenerates mitochondria via Arl2 in neonatal rat cardiac myocytes. J. Biol. Chem. 285, 4920–4930. doi: 10.1074/jbc.m109.082610
Howald, H., and Hoppeler, H. (2003). Performing at extreme altitude: muscle cellular and subcellular adaptations. Eur. J. Appl. Physiol. 90, 360–364. doi: 10.1007/s00421-003-0872-9
Hu, H., Du, L., Nagabayashi, G., Seeger, R. C., and Gatti, R. A. (2010). ATM is down-regulated by N-Myc–regulated microRNA-421. Proc. Natl. Acad. Sci. U.S. A. 107, 1506–1511. doi: 10.1073/pnas.0907763107
Huang, X., Ding, L., Bennewith, K. L., Tong, R. T., Welford, S. M., Ang, K. K., et al. (2009). Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol. Cell. 35, 856–867. doi: 10.1016/j.molcel.2009.09.006
Huang, X., Le, Q. T., and Giaccia, A. J. (2010). MiR-210–micromanager of the hypoxia pathway. Trends Mol. Med. 16, 230–237. doi: 10.1016/j.molmed.2010.03.004
Incoronato, M., Garofalo, M., Urso, L., Romano, G., Quintavalle, C., Zanca, C., et al. (2010). miR-212 increases tumor necrosis factor-related apoptosis-inducing ligand sensitivity in non-small cell lung cancer by targeting the antiapoptotic protein PED. Cancer Res. 70, 3638–3646. doi: 10.1158/0008-5472.can-09-3341
Ivan, M., Kondo, K., Yang, H., Kim, W., Valiando, J., Ohh, M., et al. (2001). HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468. doi: 10.1126/science.1059817
Jaakkola, P., Mole, D. R., Tian, Y.-M., Wilson, M. I., Gielbert, J., Gaskell, S. J., et al. (2001). Targeting of HIF-α to the von hippel-lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472. doi: 10.1126/science.1059796
Jarmakani, J. M., Nagatomo, T., Nakazawa, M., and Langer, G. A. (1978). Effect of hypoxia on myocardial high-energy phosphates in the neonatal mammalian heart. Am. J. Physiol. 235, H475–H481. doi: 10.1152/ajpheart.1978.235.5.H475
Jason, M., Caitlin, R., Ping, C., and Burge, C. B. (2012). Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science 338, 1593–1599. doi: 10.1126/science.1228186
Jin, W., Dodson, M. V., Moore, S. S., Basarab, J. A., and Guan, L. L. (2010). Characterization of microRNA expression in bovine adipose tissues: a potential regulatory mechanism of subcutaneous adipose tissue development. BMC Mol. Biol. 11:29. doi: 10.1186/1471-2199-11-29
Kam, H., Ou, L., Thron, C. D., Smith, R., and Leiter, J. C. (1999). Role of the spleen in the exaggerated polycythemic response to hypoxia in chronic mountain sickness in rats. J. Appl. Physiol. 87, 1901–1908. doi: 10.1152/jappl.1999.87.5.1901
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Komolova, G. S., and Egorov, I. A. (1985). Characteristics of the transcription activity of liver nuclear DNA in long-term adaptation to altitude hypoxia. Izv. Akad. Nauk SSSR Biol. 1, 25–30.
Kozomara, A., and Griffiths-Jones, S. (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73. doi: 10.1093/nar/gkt1181
Kuhnert, F., Mancuso, M. R., Hampton, J., Stankunas, K., Asano, T., Chen, C. Z., et al. (2008). Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development 135, 3989–3993. doi: 10.1242/dev.029736
Kulshreshtha, R., Ferracin, M., Wojcik, S. E., Garzon, R., Alder, H., Agosto-Perez, F. J., et al. (2007). A microRNA signature of hypoxia. Mol. Cell Biol. 27, 1859–1867. doi: 10.1128/MCB.01395-06
Langmead, B. (2010). Aligning short sequencing reads with bowtie. Curr. Protoc. Bioinformatics 32, 11–17. doi: 10.1002/0471250953.bi1107s32
Levett, D. Z., Radford, E. J., Menassa, D. A., Franziska, E. G., Morash, A. J., Hans, H., et al. (2012). Acclimatization of skeletal muscle mitochondria to high-altitude hypoxia during an ascent of Everest. FASEB J. 26, 1431–1432.
Levine, A. J., Hu, W., and Feng, Z. (2006). The P53 pathway: what questions remain to be explored? Cell Death Differ. 13, 1027–1036. doi: 10.1038/sj.cdd.4401910
Li, S., Zheng, B., Wang, Y., Yan, C., Jian, T., and Xun, L. (2010). Significance of changes of blood examination, arterial blood gas and blood biochemistry in early diagnosis of high altitude pulmonary edema. Med. J. National Defend. Forces Southwest China 20, 415–417.
Li, W. H., Yuan, D. Y., Sun, F. Y., Zhao, F. C., Zhang, M., and Liu, Z. (2012). The effects of high altitude hypoxia environment on ultrastructure and the expression of HIF-1α in the rat lung. Appl. Mech. Mater. 140, 68–73. doi: 10.4028/www.scientific.net/amm.140.68
Li, W. Q., Chen, C., Xu, M. D., Guo, J., Li, Y. M., Xia, Q. M., et al. (2011). The rno-miR-34 family is upregulated and targets ACSL1 in dimethylnitrosamine-induced hepatic fibrosis in rats. FEBS J. 278, 1522–1532. doi: 10.1111/j.1742-4658.2011.08075.x
Lin, Q., Gao, Z., Alarcon, R. M., Ye, J., and Yun, Z. (2009). A role of miR-27 in the regulation of adipogenesis. FEBS J. 276, 2348–2358.
Lin, X., Dexin, Z., Rui, D., Yanglin, P., Lina, Z., Shiren, S., et al. (2010). miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int. J. Cancer 123, 372–379. doi: 10.1002/ijc.23501
Liu, P., Hu, Y., Ma, L., Du, M., Xia, L., and Hu, Z. (2015). miR-425 inhibits melanoma metastasis through repression of PI3K-Akt pathway by targeting IGF-1. Biomed. Pharmacother. 75, 51–57. doi: 10.1016/j.biopha.2015.08.010
Liu, Z. (2011). Lung tissue ultrastructure and hypoxia-inducible factor 1 alpha expression in the rats exposed to high altitude hypoxia. J. Clin. Rehab. Tissue Eng. Res. 15, 6905–6908.
Lovis, P., Gattesco, S., and Regazzi, R. (2008a). Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol. Chem. 389, 305–312. doi: 10.1515/BC.2008.026
Lovis, P., Roggli, E., Laybutt, D. R., Gattesco, S., Yang, J. Y., Widmann, C., et al. (2008b). Alterations in microRNA expression contribute to fatty acid-induced pancreatic beta-cell dysfunction. Diabetes 57, 2728–2736. doi: 10.2337/db07-1252
Lu, J., Guo, S., Ebert, B. L., Zhang, H., Peng, X., Bosco, J., et al. (2008). MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev. Cell. 14, 843–853. doi: 10.1016/j.devcel.2008.03.012
Ludwig, N., Leidinger, P., Becker, K., Backes, C., Fehlmann, T., Pallasch, C., et al. (2016). Distribution of miRNA expression across human tissues. Nucleic Acids Res. 44, 3865–3877. doi: 10.1093/nar/gkw116
Luks, A. M., Johnson, R. J., and Swenson, E. R. (2008). Chronic kidney disease at high altitude. J. Am. Soc. Nephrol. 19, 2262–2271. doi: 10.1681/asn.2007111199
Lundby, C., Calbet, J. A. L., and Robach, P. (2009). The response of human skeletal muscle tissue to hypoxia. Cell. Mol. Life Sci. 66, 3615–3623. doi: 10.1007/s00018-009-0146-8
Lundby, C., Pilegaard, H., Van, H. G., Sander, M., Calbet, J., Loft, S., et al. (2003). Oxidative DNA damage and repair in skeletal muscle of humans exposed to high-altitude hypoxia. Toxicology 192, 229–236. doi: 10.1016/s0300-483x(03)00328-7
Mamatov, N., KangeldiYeva, G., and Comba, B. (2012). High altitude grassland use and research of Yaks ethology. Yüzüncü Yıl Üniversitesi Veteriner Fakültesi Dergisi 23, 41–43.
Mccarthy, D. J., Yunshun, C., and Smyth, G. K. (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297. doi: 10.1093/nar/gks042
Mihaela, P., Pertea, G. M., Antonescu, C. M., Tsung-Cheng, C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Miles, B. D., Peter, B., Michael, K., and Baumgartner, R. W. (2009). Emerging concepts in acute mountain sickness and high-altitude cerebral edema: from the molecular to the morphological. Cell. Mol. Life Sci. 66, 3583–3594. doi: 10.1007/s00018-009-0145-9
Mingzhou, L., Shilin, T., Long, J., Guangyu, Z., Ying, L., Yuan, Z., et al. (2013). Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat. Genet. 45, 1431–1438. doi: 10.1038/ng.2811
Mohanraj, P., Merola, A. J., Wright, V. P., and Clanton, T. L. (1998). Antioxidants protect rat diaphragmatic muscle function under hypoxic conditions. J. Appl. Physiol. 84, 1960–1966. doi: 10.1152/jappl.1998.84.6.1960
Moromisato, D. Y., Moromisato, M. Y., Zanconato, S., and Roberts, C. T. Jr. (1996). Effect of hypoxia on lung, heart, and liver insulin-like growth factor-I gene and receptor expression in the newborn rat. Crit. Care Med. 24, 919–924. doi: 10.1097/00003246-199606000-00008
Moskwa, P., Buffa, F. M., Pan, Y., Panchakshari, R., Gottipati, P., Muschel, R. J., et al. (2011). miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol. Cell 41, 210–220. doi: 10.1016/j.molcel.2010.12.005
Murray, A. J. (2009). Metabolic adaptation of skeletal muscle to high altitude hypoxia: How new technologies could resolve the controversies. Genome Med. 1:117. doi: 10.1186/gm117
Naeije, R. (2010). Physiological adaptation of the cardiovascular system to high altitude. Prog. Cardiovasc. Dis. 52, 456–466. doi: 10.1016/j.pcad.2010.03.004
Najafi-Shoushtari, S. H., Kristo, F., Li, Y., Shioda, T., Cohen, D. E., Gerszten, R. E., et al. (2010). MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 328, 1566–1569. doi: 10.1126/science.1189123
Nakanishi, N., Nakagawa, Y., Tokushige, N., Aoki, N., Matsuzaka, T., Ishii, K., et al. (2009). The up-regulation of microRNA-335 is associated with lipid metabolism in liver and white adipose tissue of genetically obese mice. Biochem. Biophys. Res. Commun. 385, 492–496. doi: 10.1016/j.bbrc.2009.05.058
Nakayama, K., Qi, J., and Ronai, Z. (2009). The ubiquitin ligase siah2 and the hypoxia response. Mol. Cancer Res. 7, 443–451. doi: 10.1158/1541-7786.mcr-08-0458
Newgard, C. B., Hwang, P. K., and Fletterick, R. J. (1989). The family of glycogen phosphorylases: structure and function. Crit. Rev. Biochem. 24, 69–99.
Ou, L. C., Kim, D., Layton, W. M. Jr., and Smith, R. P. (1980). Splenic erythropoiesis in polycythemic response of the rat to high-altitude exposure. J. Appl. Physiol. Respir. Environ. Exerc. Physiol. 48, 857–861. doi: 10.1152/jappl.1980.48.5.857
Pandey, A. K., Verma, G., Vig, S., Srivastava, S., Srivastava, A. K., and Datta, M. (2011). miR-29a levels are elevated in the db/db mice liver and its overexpression leads to attenuation of insulin action on PEPCK gene expression in HepG2 cells. Mol. Cell Endocrinol. 332, 125–133. doi: 10.1016/j.mce.2010.10.004
Papagiannakopoulos, T., Shapiro, A., and Kosik, K. S. (2008). MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 68, 8164–8172. doi: 10.1158/0008-5472.can-08-1305
Park, A. M., Nagase, H., Vinod Kumar, S., and Suzuki, Y. J. (2007). Acute intermittent hypoxia activates myocardial cell survival signaling. Am. J. Physiol. Heart Circ. Physiol. 292, H751–H757. doi: 10.1152/ajpheart.01016.2006
Pierson, J., Hostager, B., Fan, R., and Vibhakar, R. (2008). Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J. Neurooncol. 90, 1–7. doi: 10.1007/s11060-008-9624-3
Pietenpol, J. A., and Stewart, Z. A. (2002). Cell cycle checkpoint signaling: : cell cycle arrest versus apoptosis. Toxicology 181, 475–481.
Pilotte, J., Dupont-Versteegden, E. E., and Vanderklish, P. W. (2011). Widespread regulation of miRNA biogenesis at the Dicer step by the cold-inducible RNA-binding protein. RBM3. PLoS One 6:e28446. doi: 10.1371/journal.pone.0028446
Plaisance, V., Abderrahmani, A., Perret-Menoud, V., Jacquemin, P., Lemaigre, F., and Regazzi, R. (2006). MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J. Biol. Chem. 281, 26932–26942. doi: 10.1074/jbc.M601225200
Pocock, R. (2011). Invited review: decoding the microRNA response to hypoxia. Pflugers Arch. 461, 307–315. doi: 10.1007/s00424-010-0910-5
Pothof, J., Verkaik, N. S., van, I. W., Wiemer, E. A., Ta, V. T., van der Horst, G. T., et al. (2009). MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. EMBO J. 28, 2090–2099. doi: 10.1038/emboj.2009.156
Poy, M. N., Eliasson, L., Krutzfeldt, J., Kuwajima, S., Ma, X., Macdonald, P. E., et al. (2004). A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432, 226–230. doi: 10.1038/nature03076
Poy, M. N., Hausser, J., Trajkovski, M., Braun, M., Collins, S., Rorsman, P., et al. (2009). miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc. Natl. Acad. Sci. U.S.A. 106, 5813–5818. doi: 10.1073/pnas.0810550106
Qu, Y., Zhao, H., Han, N., Zhou, G., Song, G., Gao, B., et al. (2013). Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat. Commun. 4:2071. doi: 10.1038/ncomms3071
Rayner, K. J., Sheedy, F. J., Esau, C. C., Hussain, F. N., Temel, R. E., Parathath, S., et al. (2011). Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J. Clin. Invest. 121, 2921–2931. doi: 10.1172/JCI57275
Rayner, K. J., Suarez, Y., Davalos, A., Parathath, S., Fitzgerald, M. L., Tamehiro, N., et al. (2010). MiR-33 contributes to the regulation of cholesterol homeostasis. Science 328, 1570–1573. doi: 10.1126/science.1189862
Richalet, J. P. (2007). A proposed classification of environmental adaptation: the example of high altitude. Rev. Environ. Sci. Biotechnol. 6, 223–229. doi: 10.1007/s11157-006-9113-0
Richardson, M. X., Lodin, A., Reimers, J., and Schagatay, E. (2008). Short-term effects of normobaric hypoxia on the human spleen. Eur. J. Appl. Physiol. 104, 395–399. doi: 10.1007/s00421-007-0623-4
Robinson, M. D., Mccarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140.
Rodriguez-Manzaneque, J. C., Lane, T. F., Ortega, M. A., Hynes, R. O., Lawler, J., and Iruela-Arispe, M. L. (2001). Thrombospondin-1 suppresses spontaneous tumor growth and inhibits activation of matrix metalloproteinase-9 and mobilization of vascular endothelial growth factor. Proc. Natl. Acad. Sci. U.S.A. 98, 12485–12490. doi: 10.1073/pnas.171460498
Rong, L., and Gastroenterology, D. O. (2009). High-altitude hypoxia and liver injury. World Chin. J. Digestol. 17, 2171–2178.
Saltiel, A. R., and Kahn, C. R. (2001). Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414, 799–806. doi: 10.1038/414799a
Schickel, R., Park, S. M., Murmann, A. E., and Peter, M. E. (2010). mir-200c regulates induction of apoptosis through CD95 by Targeting FAP-1. Mol. Cell 38, 908–915. doi: 10.1016/j.molcel.2010.05.018
Scott, G. R., Schulte, P. M., Egginton, S., Scott, A. L., Richards, J. G., and Milsom, W. K. (2011). Molecular evolution of cytochrome C oxidase underlies high-altitude adaptation in the bar-headed goose. Mol. Biol. Evol. 28, 351–363. doi: 10.1093/molbev/msq205
Semenza, G. L. (2007). Hypoxia-inducible factor 1 (HIF-1) pathway. Sci. STKE 2007:cm8. doi: 10.1126/stke.4072007cm8
Shibuya, M. (2006). Vascular endothelial growth factor receptor-1 (VEGFR-1/Flt-1): a dual regulator for angiogenesis. Angiogenesis 9, 225–230. doi: 10.1007/s10456-006-9055-8
Silvia, G., Neri, M., Ezio, G., Simone, M., Giovanni Vanni, F., Silvia Anna, C., et al. (2007). miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J. Biol. Chem. 282, 23716–23724. doi: 10.1074/jbc.m701805200
Simonson, T. S., Yang, Y., Huff, C. D., Yun, H., Qin, G., Witherspoon, D. J., et al. (2010). Genetic evidence for high-altitude adaptation in Tibet. Science 329, 72–75. doi: 10.1126/science.1189406
Skrzypczakjankun, E. (2001). Recombinant PAI-1 inhibits angiogenesis and reduces size of LNCaP prostate cancer xenografts in SCID mice. Oncol. Rep. 8, 463–470.
Small, E. M., Frost, R. J. A., and Olson, E. N. (2010). MicroRNAs add a new dimension to cardiovascular disease. Circulation 121:1022. doi: 10.1161/circulationaha.109.889048
Storz, J. F., Runck, A. M., Sabatino, S. J., Kelly, J. K., Nuno, F., Hideaki, M., et al. (2009). Evolutionary and functional insights into the mechanism underlying high-altitude adaptation of deer mouse hemoglobin. Proc. Natl. Acad. Sci. U.S.A. 106, 14450–14455. doi: 10.1073/pnas.0905224106
Sun, T., Fu, M., Bookout, A. L., Kliewer, S. A., and Mangelsdorf, D. J. (2009). MicroRNA let-7 regulates 3T3-L1 adipogenesis. Mol. Endocrinol. 23, 925–931. doi: 10.1210/me.2008-0298
Swulius, M. T., and Waxham, M. N. (2008). Ca 2+ /calmodulin-dependent protein kinases. Cell. Mol. Life Sci. 65, 2637–2657.
Takanabe, R., Ono, K., Abe, Y., Takaya, T., Horie, T., Wada, H., et al. (2008). Up-regulated expression of microRNA-143 in association with obesity in adipose tissue of mice fed high-fat diet. Biochem. Biophys. Res. Commun. 376, 728–732. doi: 10.1016/j.bbrc.2008.09.050
Tang, Q., Huang, W., Guan, J., Jin, L., Che, T., Fu, Y., et al. (2015). Transcriptomic analysis provides insight into high-altitude acclimation in domestic goats. Gene 567, 208–216. doi: 10.1016/j.gene.2015.05.007
Trajkovski, M., Hausser, J., Soutschek, J., Bhat, B., Akin, A., Zavolan, M., et al. (2011). MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 474, 649–653. doi: 10.1038/nature10112
Tung, H. Y. L., Alemany, S., and Cohen, P. (1984). The protein phosphatases involved in cellular regulation. Febs. J. 145, 305–313.
Wan, G., Mathur, R., Hu, X., Zhang, X., and Lu, X. (2011). miRNA response to DNA damage. Trends Biochem. Sci. 36, 478–484. doi: 10.1016/j.tibs.2011.06.002
Wan, P., Zou, F., Zhang, X., Li, H., Dulak, A., Tomko R. J., Jr., et al. (2009). MicroRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res. 69, 8157–8165. doi: 10.1158/0008-5472.can-09-1996
Wang, G. D., Fan, R. X., Zhai, W., Liu, F., Wang, L., Zhong, L., et al. (2014). Genetic convergence in the adaptation of dogs and humans to the high-altitude environment of the tibetan plateau. Genome Biol. Evol. 6:2122. doi: 10.1093/gbe/evu162
Wang, Q., Huang, Z., Xue, H., Jin, C., Ju, X. L., Han, J. D., et al. (2008a). MicroRNA miR-24 inhibits erythropoiesis by targeting activin type I receptor ALK4. Blood 111, 588–595. doi: 10.1182/blood-2007-05-092718
Wang, S., Aurora, A. B., Johnson, B. A., Qi, X., McAnally, J., Hill, J. A., et al. (2008b). The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 15, 261–271. doi: 10.1016/j.devcel.2008.07.002
Wang, R., Zhang, P., Li, J., Guan, H., and Shi, G. (2016). Ubiquitination is absolutely required for the degradation of hypoxia-inducible factor - 1 alpha protein in hypoxic conditions. Biochem. Biophys. Res. Commun. 470, 117–122. doi: 10.1016/j.bbrc.2016.01.005
Wang, Y., Li, Z., Xu, P., Huang, L., Tong, J., Huang, H., et al. (2011). Angiomotin-like2 gene (amotl2) is required for migration and proliferation of endothelial cells during angiogenesis. J. Biol. Chem. 286, 41095–41104. doi: 10.1074/jbc.m111.296806
Wei, F., Liu, Y., Guo, Y., Xiang, A., Wang, G., Xue, X., et al. (2013). miR-99b-targeted mTOR induction contributes to irradiation resistance in;pancreatic cancer. Mol. Cancer 12:81. doi: 10.1186/1476-4598-12-81
Wichern, D. W., and Johnson, R. A. (1982). Applied Multivariate Statistical Analysis. London: Pearson.
Wilson, A. T., Reilly, C. S., Davies, S. W., and Wedzicha, J. A. (1993). Hypoxia and the heart. Br. Heart J. 70:98. doi: 10.1136/hrt.70.1.98-a
Wurdinger, T., Tannous, B. A., Saydam, O., Skog, J., Grau, S., Soutschek, J., et al. (2008). miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell 14, 382–393. doi: 10.1016/j.ccr.2008.10.005
Xie, H., Lim, B., and Lodish, H. F. (2009). MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes 58, 1050–1057. doi: 10.2337/db08-1299
Yang, J., Jin, Z. B., Chen, J., Huang, X. F., Li, X. M., Liang, Y. B., et al. (2017). Genetic signatures of high-altitude adaptation in Tibetans. Proc. Natl. Acad. Sci. U.S.A. 114, 4189–4194. doi: 10.1073/pnas.1617042114
Yang, X., Feng, M., Jiang, X., Wu, Z., Li, Z., Aau, M., et al. (2009). miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb–E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev. 23, 2388–2393. doi: 10.1101/gad.1819009
Yijiang, Z., Jianhua, Z., and Feili, L. (2013). Acute kidney injury at high altitude. High Alt. Med. Biol. 14, 183–185. doi: 10.1089/ham.2012.1123
Ying-Jie, P., Guoxiang, Y., Deviprasadh, R., Sharma, S. D., Marta, B. M., Kumar, G. K., et al. (2010). Heterozygous HIF-1alpha deficiency impairs carotid body-mediated systemic responses and reactive oxygen species generation in mice exposed to intermittent hypoxia. J. Physiol. 577, 705–716. doi: 10.1113/jphysiol.2006.114033
Yu, Y., Wang, Y., Ren, X., Tsuyada, A., Li, A., Liu, L. J., et al. (2010). Context-dependent bidirectional regulation of the MutS homolog 2 by transforming growth factor β contributes to chemoresistance in breast cancer cells. Mol. Cancer Res. 8, 1633–1642. doi: 10.1158/1541-7786.mcr-10-0362
Yuan, G., Nanduri, J. S., Semenza, G., and Prabhakar, N. (2010). Induction of HIF-1alpha expression by intermittent hypoxia: involvement of NADPH oxidase, Ca2+ signaling, prolyl hydroxylases, and mTOR. J. Cell. Physiol. 217, 674–685. doi: 10.1002/jcp.21537
Yuan, J. Y., Wang, F., Yu, J., Yang, G. H., Liu, X. L., and Zhang, J. W. (2009). MicroRNA-223 reversibly regulates erythroid and megakaryocytic differentiation of K562 cells. J. Cell Mol. Med. 13, 4551–4559. doi: 10.1111/j.1582-4934.2008.00585.x
Zhang, J., Chen, L., Long, K. R., and Mu, Z. P. (2015). Hypoxia-related gene expression in porcine skeletal muscle tissues at different altitude. Genet. Mol. Res. 14, 11587–11593. doi: 10.4238/2015.september.28.10
Zhang, X., Wan, G., Mlotshwa, S., Vance, V., Berger, F. G., Chen, H., et al. (2010). Oncogenic wip1 phosphatase is inhibited by miR-16 in the DNA damage signaling pathway. Cancer Res. 70, 7176–7186. doi: 10.1158/0008-5472.can-10-0697
Zhao, J. J., Lin, J., Lwin, T., Yang, H., Guo, J., Kong, W., et al. (2010). microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood 115, 2630–2639. doi: 10.1182/blood-2009-09-243147
Zhou, X., Wang, B., Pan, Q., Zhang, J., Kumar, S., Sun, X., et al. (2014). Whole-genome sequencing of the snub-nosed monkey provides insights into folivory and evolutionary history. Nat. Genet. 46, 1303–1310. doi: 10.1038/ng.3137
Keywords: miRNA, high-altitude acclimatization, goat, multiple tissue, angiogenesis
Citation: Feng S, Ma J, Long K, Zhang J, Qiu W, Li Y, Jin L, Wang X, Jiang A, Liu L, Xiao W, Li X, Tang Q and Li M (2020) Comparative microRNA Transcriptomes in Domestic Goats Reveal Acclimatization to High Altitude. Front. Genet. 11:809. doi: 10.3389/fgene.2020.00809
Received: 09 February 2020; Accepted: 06 July 2020;
Published: 31 July 2020.
Edited by:
Peng Xu, Xiamen University, ChinaReviewed by:
Huanling Wang, Huazhong Agricultural University, ChinaTao Zhou, Auburn University, United States
Copyright © 2020 Feng, Ma, Long, Zhang, Qiu, Li, Jin, Wang, Jiang, Liu, Xiao, Li, Tang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qianzi Tang, wupie@163.com; Mingzhou Li, mingzhou.li@sicau.edu.cn
†These authors have contributed equally to this work
 Siyuan Feng†
Siyuan Feng† Long Jin
Long Jin Mingzhou Li
Mingzhou Li