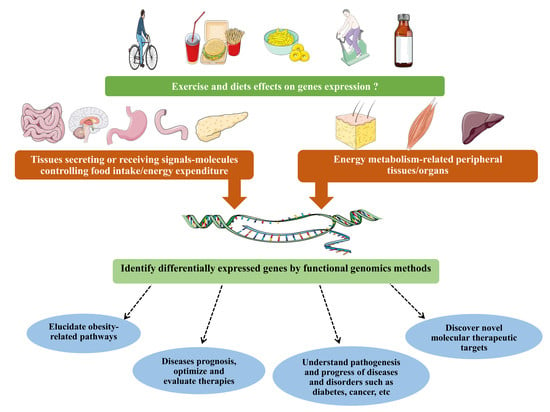
Exercise and High-Fat Diet in Obesity: Functional Genomics Perspectives of Two Energy Homeostasis Pillars
Abstract
:1. Obesity as a Health Problem in Need of Novel Approaches
2. Exercise-Related Genes and Pathways: Towards an Exercise Pill
2.1. Exercise and Health
2.2. Exercise Impacts Gene Expression
2.3. Gene Expression Patterns Underlie Muscular Adaptation to Exercise
2.4. Implications
3. Diet-Related Genes: A Focus on High-Fat Diet to Identify a Lipid-Specific Signal
3.1. High-Fat Diet Particularities in Obesity Context
3.2. Digestive System (First Food “Receptors”)
3.3. Adipose Tissue (Energy-Stocking Tissue) and Skeletal Muscle (Energy-Usage Tissue)
3.4. Brain (Energy Balance-Control Centers)
3.5. Potential Applications
4. Conclusions, Discussion, and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Obesity: Preventing and managing the global epidemic. Report of a WHO consultation. In WHO Technical Report Series 894; World Health Organization: Geneva, Switzerland, 2000; pp. i-xii, 1-253. [Google Scholar]
- Ghanemi, A.; Yoshioka, M.; St-Amand, J. Broken Energy Homeostasis and Obesity Pathogenesis: The Surrounding Concepts. J. Clin. Med. 2018, 7, 453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, Y.; Qin, B.; Poti, J.; Sokol, R.; Gordon-Larsen, P. Epidemiology of Obesity in Adults: Latest Trends. Curr. Obes. Rep. 2018, 7, 276–288. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.; Fleming, T.; Robinson, M.; Thomson, B.; Graetz, N.; Margono, C.; Mullany, E.C.; Biryukov, S.; Abbafati, C.; Abera, S.F.; et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet 2014, 384, 766–781. [Google Scholar] [CrossRef] [Green Version]
- Ghanemi, A.; Yoshioka, M.; St-Amand, J. Will an obesity pandemic replace the coronavirus disease-2019 (COVID-19) pandemic? Med. Hypotheses 2020, 144, 110042. [Google Scholar] [CrossRef]
- Andolfi, C.; Fisichella, P.M. Epidemiology of Obesity and Associated Comorbidities. J. Laparoendosc. Adv. Surg. Tech. A 2018, 28, 919–924. [Google Scholar] [CrossRef]
- Bhupathiraju, S.N.; Hu, F.B. Epidemiology of Obesity and Diabetes and Their Cardiovascular Complications. Circ. Res. 2016, 118, 1723–1735. [Google Scholar] [CrossRef]
- Wang, J.; Wu, Y.; Xiong, G.; Chao, T.; Jin, Q.; Liu, R.; Hao, L.; Wei, S.; Yang, N.; Yang, X. Introduction of complementary feeding before 4 months of age increases the risk of childhood overweight or obesity: A meta-analysis of prospective cohort studies. Nutr. Res. 2016, 36, 759–770. [Google Scholar] [CrossRef]
- Segal, M.; Eliasziw, M.; Phillips, S.; Bandini, L.; Curtin, C.; Kral, T.V.E.; Sherwood, N.E.; Sikich, L.; Stanish, H.; Must, A. Intellectual disability is associated with increased risk for obesity in a nationally representative sample of U.S. children. Disabil. Health J. 2016, 9, 392–398. [Google Scholar] [CrossRef] [Green Version]
- Michopoulos, V. Stress-Induced alterations in estradiol sensitivity increase risk for obesity in women. Physiol. Behav. 2016. [Google Scholar] [CrossRef] [Green Version]
- Heymsfield, S.B.; Wadden, T.A. Mechanisms, Pathophysiology, and Management of Obesity. N. Engl. J. Med. 2017, 376, 254–266. [Google Scholar] [CrossRef]
- Kim, J.Y.; Park, D.I.; Yu, J.; Jung, Y.S.; Park, J.H.; Kim, H.J.; Cho, Y.K.; Sohn, C.I.; Jeon, W.K.; Kim, B.I.; et al. Increased Risk of Advanced Colorectal Neoplasia Among Korean Men With Metabolic Abnormality and Obesity. Clin. Gastroenterol. Hepatol. 2016, 14. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.C.; D′Ambrosia, C.; McLawhorn, A.S.; Schairer, W.W.; Padgett, D.E.; Cross, M.B. Malnutrition Increases With Obesity and Is a Stronger Independent Risk Factor for Postoperative Complications: A Propensity-Adjusted Analysis of Total Hip Arthroplasty Patients. J. Arthroplast. 2016. [Google Scholar] [CrossRef] [PubMed]
- Ricketts, T.A.; Sui, X.; Lavie, C.J.; Blair, S.N.; Ross, R. Addition of Cardiorespiratory Fitness Within an Obesity Risk Classification Model Identifies Men at Increased Risk of All-Cause Mortality. Am. J. Med. 2016, 129, 536.e13–536.e20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontaine, K.R.; Redden, D.T.; Wang, C.; Westfall, A.O.; Allison, D.B. Years of life lost due to obesity. JAMA 2003, 289, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Ghanemi, A.; St-Amand, J. Redefining obesity toward classifying as a disease. Eur. J. Intern. Med. 2018, 55, 20–22. [Google Scholar] [CrossRef] [PubMed]
- le Roux, C.W.; Heneghan, H.M. Bariatric Surgery for Obesity. Med. Clin. N. Am. 2018, 102, 165–182. [Google Scholar] [CrossRef]
- Nuffer, W. Chapter 5-Pharmacologic Agents Chapter for Abdominal Obesity. In Nutrition in the Prevention and Treatment of Abdominal Obesity, 2nd ed.; Watson, R.R., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 51–66. [Google Scholar] [CrossRef]
- Kim, G.W.; Lin, J.E.; Blomain, E.S.; Waldman, S.A. Antiobesity pharmacotherapy: New drugs and emerging targets. Clin. Pharmacol. Ther. 2014, 95, 53–66. [Google Scholar] [CrossRef]
- Gadde, K.M.; Pritham Raj, Y. Pharmacotherapy of Obesity: Clinical Trials to Clinical Practice. Curr. Diab. Rep. 2017, 17, 34. [Google Scholar] [CrossRef]
- Gadde, K.M.; Apolzan, J.W.; Berthoud, H.R. Pharmacotherapy for Patients with Obesity. Clin. Chem. 2017. [Google Scholar] [CrossRef] [Green Version]
- Canella, D.S.; Novaes, H.M.; Levy, R.B. Medicine expenses and obesity in Brazil: An analysis based on the household budget survey. BMC Public Health 2016, 16, 54. [Google Scholar] [CrossRef] [Green Version]
- Verhaeghe, N.; De Greve, O.; Annemans, L. The potential health and economic effect of a Body Mass Index decrease in the overweight and obese population in Belgium. Public Health 2016. [Google Scholar] [CrossRef] [PubMed]
- Westerterp, K.R. Control of Energy Expenditure in Humans. In Endotext; Feingold, K.R., Anawalt, B., Boyce, A., Chrousos, G., Dungan, K., Grossman, A., Hershman, J.M., Kaltsas, G., Koch, C., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Galloza, J.; Castillo, B.; Micheo, W. Benefits of Exercise in the Older Population. Phys. Med. Rehabil. Clin. N Am. 2017, 28, 659–669. [Google Scholar] [CrossRef] [PubMed]
- Myers, J. Cardiology patient pages. Exercise and cardiovascular health. Circulation 2003, 107, e2–e5. [Google Scholar] [CrossRef] [Green Version]
- Tran, Z.V.; Weltman, A. Differential effects of exercise on serum lipid and lipoprotein levels seen with changes in body weight. A meta-Analysis. Jama 1985, 254, 919–924. [Google Scholar] [CrossRef] [PubMed]
- Blundell, J.E.; Gibbons, C.; Caudwell, P.; Finlayson, G.; Hopkins, M. Appetite control and energy balance: Impact of exercise. Obes. Rev. 2015, 16 (Suppl. 1), 67–76. [Google Scholar] [CrossRef] [Green Version]
- Chin, S.H.; Kahathuduwa, C.N.; Binks, M. Physical activity and obesity: What we know and what we need to know. Obes. Rev. 2016, 17, 1226–1244. [Google Scholar] [CrossRef] [PubMed]
- Marcin, T.; Eser, P.; Prescott, E.; Mikkelsen, N.; Prins, L.F.; Kolkman, E.K.; Lado-Baleato, Ó.; Cardaso-Suaréz, C.; Bruins, W.; van der Velde, A.E.; et al. Predictors of pre-Rehabilitation exercise capacity in elderly European cardiac patients-The EU-CaRE study. Eur. J. Prev. Cardiol. 2019. [Google Scholar] [CrossRef]
- McLeod, M.; Breen, L.; Hamilton, D.L.; Philp, A. Live strong and prosper: The importance of skeletal muscle strength for healthy ageing. Biogerontology 2016, 17, 497–510. [Google Scholar] [CrossRef] [Green Version]
- Brook, M.S.; Wilkinson, D.J.; Phillips, B.E.; Perez-Schindler, J.; Philp, A.; Smith, K.; Atherton, P.J. Skeletal muscle homeostasis and plasticity in youth and ageing: Impact of nutrition and exercise. Acta Physiol. (Oxf.) 2016, 216, 15–41. [Google Scholar] [CrossRef] [Green Version]
- Shad, B.J.; Wallis, G.; van Loon, L.J.; Thompson, J.L. Exercise prescription for the older population: The interactions between physical activity, sedentary time, and adequate nutrition in maintaining musculoskeletal health. Maturitas 2016, 93, 78–82. [Google Scholar] [CrossRef]
- Figueira, A.C.C.; Figueira, M.C.; Silva, C.; Padrao, A.; Oliveira, P.A.; Ferreira, R.P.; Duarte, J.A. Exercise Training-Induced Modulation in Microenvironment of Rat Mammary Neoplasms. Int. J. Sports Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, L.; Idorn, M.; Olofsson, G.H.; Lauenborg, B.; Nookaew, I.; Hansen, R.H.; Johannesen, H.H.; Becker, J.C.; Pedersen, K.S.; Dethlefsen, C.; et al. Voluntary Running Suppresses Tumor Growth through Epinephrine- and IL-6-Dependent NK Cell Mobilization and Redistribution. Cell Metab. 2016, 23, 554–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antunes, J.M.M.; Ferreira, R.M.P.; Moreira-Goncalves, D. Exercise Training as Therapy for Cancer-Induced Cardiac Cachexia. Trends Mol. Med. 2018, 24, 709–727. [Google Scholar] [CrossRef] [PubMed]
- Wesnes, K.; Myhr, K.M.; Riise, T.; Cortese, M.; Pugliatti, M.; Bostrom, I.; Landtblom, A.M.; Wolfson, C.; Bjornevik, K. Physical activity is associated with a decreased multiple sclerosis risk: The EnvIMS study. Mult. Scler. 2018, 24, 150–157. [Google Scholar] [CrossRef]
- Han, P.; Zhang, W.; Kang, L.; Ma, Y.; Fu, L.; Jia, L.; Yu, H.; Chen, X.; Hou, L.; Wang, L.; et al. Clinical Evidence of Exercise Benefits for Stroke. Adv. Exp. Med. Biol. 2017, 1000, 131–151. [Google Scholar] [CrossRef]
- Panchik, D.; Masco, S.; Zinnikas, P.; Hillriegel, B.; Lauder, T.; Suttmann, E.; Chinchilli, V.; McBeth, M.; Hermann, W. The Effect of Exercise on Breast Cancer-Related Lymphedema: What the Lymphatic Surgeon Needs to Know. J. Reconstr. Microsurg. 2018. [Google Scholar] [CrossRef]
- Keilani, M.; Hasenoehrl, T.; Baumann, L.; Ristl, R.; Schwarz, M.; Marhold, M.; Sedghi Komandj, T.; Crevenna, R. Effects of resistance exercise in prostate cancer patients: A meta-analysis. Support Care Cancer 2017, 25, 2953–2968. [Google Scholar] [CrossRef] [Green Version]
- Hart, N.H.; Galvao, D.A.; Newton, R.U. Exercise medicine for advanced prostate cancer. Curr. Opin. Support Palliat. Care 2017, 11, 247–257. [Google Scholar] [CrossRef]
- Erlich, A.T.; Brownlee, D.M.; Beyfuss, K.; Hood, D.A. Exercise induces TFEB expression and activity in skeletal muscle in a PGC-1alpha-Dependent manner. Am. J. Physiol. Cell Physiol. 2017. [Google Scholar] [CrossRef]
- Vainshtein, A.; Tryon, L.D.; Pauly, M.; Hood, D.A. Role of PGC-1alpha during acute exercise-Induced autophagy and mitophagy in skeletal muscle. Am. J. Physiol. Cell Physiol. 2015, 308, C710–C719. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Li, C.G.; Qi, Z.; Cui, D.; Ding, S. Acute exercise stress promotes Ref1/Nrf2 signalling and increases mitochondrial antioxidant activity in skeletal muscle. Exp. Physiol. 2016, 101, 410–420. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Liu, X.; Li, P.; Akimoto, T.; Lee, S.Y.; Zhang, M.; Yan, Z. Transcriptional profiling in mouse skeletal muscle following a single bout of voluntary running: Evidence of increased cell proliferation. J. Appl. Physiol. (Bethesda, Md. 1985) 2005, 99, 2406–2415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.; Zhou, L.; He, T. Potential effect of exercise in ameliorating insulin resistance at transcriptome level. J. Sports Med. Phys. Fit. 2017. [Google Scholar] [CrossRef] [PubMed]
- Larose, M.; St-Amand, J.; Yoshioka, M.; Belleau, P.; Morissette, J.; Labrie, C.; Raymond, V.; Labrie, F. Transcriptome of mouse uterus by serial analysis of gene expression (SAGE): Comparison with skeletal muscle. Mol. Reprod. Dev. 2004, 68, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Vissing, K.; Schjerling, P. Simplified data access on human skeletal muscle transcriptome responses to differentiated exercise. Sci. Data 2014, 1, 140041. [Google Scholar] [CrossRef] [PubMed]
- Lindholm, M.E.; Huss, M.; Solnestam, B.W.; Kjellqvist, S.; Lundeberg, J.; Sundberg, C.J. The human skeletal muscle transcriptome: Sex differences, alternative splicing, and tissue homogeneity assessed with RNA sequencing. FASEB J. 2014, 28, 4571–4581. [Google Scholar] [CrossRef] [Green Version]
- Seaborne, R.A.; Strauss, J.; Cocks, M.; Shepherd, S.; O′Brien, T.D.; van Someren, K.A.; Bell, P.G.; Murgatroyd, C.; Morton, J.P.; Stewart, C.E.; et al. Human Skeletal Muscle Possesses an Epigenetic Memory of Hypertrophy. Sci. Rep. 2018, 8, 1898. [Google Scholar] [CrossRef]
- Lindholm, M.E.; Marabita, F.; Gomez-Cabrero, D.; Rundqvist, H.; Ekstrom, T.J.; Tegner, J.; Sundberg, C.J. An integrative analysis reveals coordinated reprogramming of the epigenome and the transcriptome in human skeletal muscle after training. Epigenetics 2014, 9, 1557–1569. [Google Scholar] [CrossRef]
- Sharples, A.P.; Stewart, C.E.; Seaborne, R.A. Does skeletal muscle have an ‘epi’-memory? The role of epigenetics in nutritional programming, metabolic disease, aging and exercise. Aging Cell 2016, 15, 603–616. [Google Scholar] [CrossRef] [Green Version]
- Gensous, N.; Bacalini, M.G.; Pirazzini, C.; Marasco, E.; Giuliani, C.; Ravaioli, F.; Mengozzi, G.; Bertarelli, C.; Palmas, M.G.; Franceschi, C.; et al. The epigenetic landscape of age-related diseases: The geroscience perspective. Biogerontology 2017, 18, 549–559. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Chen, Y.; Zhang, Y.; Zhang, H.; Wu, Y.; He, H.; Gong, L.; Zeng, F.; Shi, L. Exercise during pregnancy enhances vascular function via epigenetic repression of Ca(V)1.2 channel in offspring of hypertensive rats. Life Sci. 2019, 231, 116576. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, S.A.; Baldi, J.C.; Cutfield, W.S.; McCowan, L.; Hofman, P.L. Exercise training in pregnancy reduces offspring size without changes in maternal insulin sensitivity. J. Clin. Endocrinol. Metab. 2010, 95, 2080–2088. [Google Scholar] [CrossRef] [Green Version]
- Yoshioka, M.; Tanaka, H.; Shono, N.; Snyder, E.E.; Shindo, M.; St-Amand, J. Serial analysis of gene expression in the skeletal muscle of endurance athletes compared to sedentary men. FASEB J. 2003, 17, 1812–1819. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, M.; Tanaka, H.; Shono, N.; Shindo, M.; St-Amand, J. Gene expression profile of sprinter’s muscle. Int. J. Sports Med. 2007, 28, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Cochran, A.J.; Percival, M.E.; Tricarico, S.; Little, J.P.; Cermak, N.; Gillen, J.B.; Tarnopolsky, M.A.; Gibala, M.J. Intermittent and continuous high-intensity exercise training induce similar acute but different chronic muscle adaptations. Exp. Physiol. 2014, 99, 782–791. [Google Scholar] [CrossRef] [Green Version]
- Nishida, Y.; Tanaka, H.; Tobina, T.; Murakami, K.; Shono, N.; Shindo, M.; Ogawa, W.; Yoshioka, M.; St-Amand, J. Regulation of muscle genes by moderate exercise. Int. J. Sports Med. 2010, 31, 656–670. [Google Scholar] [CrossRef]
- Riedl, I.; Yoshioka, M.; Nishida, Y.; Tobina, T.; Paradis, R.; Shono, N.; Tanaka, H.; St-Amand, J. Regulation of skeletal muscle transcriptome in elderly men after 6 weeks of endurance training at lactate threshold intensity. Exp. Gerontol. 2010, 45, 896–903. [Google Scholar] [CrossRef]
- Timmons, J.A.; Larsson, O.; Jansson, E.; Fischer, H.; Gustafsson, T.; Greenhaff, P.L.; Ridden, J.; Rachman, J.; Peyrard-Janvid, M.; Wahlestedt, C.; et al. Human muscle gene expression responses to endurance training provide a novel perspective on Duchenne muscular dystrophy. Faseb J. 2005, 19, 750–760. [Google Scholar] [CrossRef]
- Huang, J.H.; Hood, D.A. Age-Associated mitochondrial dysfunction in skeletal muscle: Contributing factors and suggestions for long-Term interventions. IUBMB Life 2009, 61, 201–214. [Google Scholar] [CrossRef]
- Marzetti, E.; Lawler, J.M.; Hiona, A.; Manini, T.; Seo, A.Y.; Leeuwenburgh, C. Modulation of age-induced apoptotic signaling and cellular remodeling by exercise and calorie restriction in skeletal muscle. Free Radic Biol. Med. 2008, 44, 160–168. [Google Scholar] [CrossRef]
- Lourenço Dos Santos, S.; Baraibar, M.A.; Lundberg, S.; Eeg-Olofsson, O.; Larsson, L.; Friguet, B. Oxidative proteome alterations during skeletal muscle ageing. Redox Biol. 2015, 5, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Melouane, A.; Yoshioka, M.; Kanzaki, M.; St-Amand, J. Sparc, an EPS-induced gene, modulates the extracellular matrix and mitochondrial function via ILK/AMPK pathways in C2C12 cells. Life Sci. 2019, 229, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Norheim, F.; Raastad, T.; Thiede, B.; Rustan, A.C.; Drevon, C.A.; Haugen, F. Proteomic identification of secreted proteins from human skeletal muscle cells and expression in response to strength training. Am. J. Physiol. Endocrinol. Metab. 2011, 301, E1013–E1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melouane, A.; Carbonell, A.; Yoshioka, M.; Puymirat, J.; St-Amand, J. Implication of SPARC in the modulation of the extracellular matrix and mitochondrial function in muscle cells. PLoS ONE 2018, 13, e0192714. [Google Scholar] [CrossRef] [Green Version]
- Aoi, W.; Naito, Y.; Takagi, T.; Tanimura, Y.; Takanami, Y.; Kawai, Y.; Sakuma, K.; Hang, L.; Mizushima, K.; Hirai, Y.; et al. A novel myokine, secreted protein acidic and rich in cysteine (SPARC), suppresses colon tumorigenesis via regular exercise. Gut 2013, 62, 882–889. [Google Scholar] [CrossRef]
- Ghanemi, A.; Melouane, A.; Yoshioka, M.; St-Amand, J. Secreted protein acidic and rich in cysteine and bioenergetics: Extracellular matrix, adipocytes remodeling and skeletal muscle metabolism. Int. J. Biochem. Cell Biol. 2019, 117, 105627. [Google Scholar] [CrossRef]
- Ghanemi, A.; Yoshioka, M.; St-Amand, J. Secreted Protein Acidic and Rich in Cysteine: Metabolic and Homeostatic Properties beyond the Extracellular Matrix Structure. Appl. Sci. 2020, 10, 2388. [Google Scholar] [CrossRef] [Green Version]
- Sage, H.; Vernon, R.B.; Funk, S.E.; Everitt, E.A.; Angello, J. SPARC, a secreted protein associated with cellular proliferation, inhibits cell spreading in vitro and exhibits Ca+2-dependent binding to the extracellular matrix. J. Cell Biol. 1989, 109, 341–356. [Google Scholar] [CrossRef]
- Francki, A.; Motamed, K.; McClure, T.D.; Kaya, M.; Murri, C.; Blake, D.J.; Carbon, J.G.; Sage, E.H. SPARC regulates cell cycle progression in mesangial cells via its inhibition of IGF-Dependent signaling. J. Cell Biochem. 2003, 88, 802–811. [Google Scholar] [CrossRef]
- Mason, I.J.; Taylor, A.; Williams, J.G.; Sage, H.; Hogan, B.L. Evidence from molecular cloning that SPARC, a major product of mouse embryo parietal endoderm, is related to an endothelial cell ‘culture shock’ glycoprotein of Mr 43,000. Embo J. 1986, 5, 1465–1472. [Google Scholar] [CrossRef]
- Cho, W.J.; Kim, E.J.; Lee, S.J.; Kim, H.D.; Shin, H.J.; Lim, W.K. Involvement of SPARC in in vitro differentiation of skeletal myoblasts. Biochem. Biophys. Res. Commun. 2000, 271, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Ghanemi, A.; Yoshioka, M.; St-Amand, J. Secreted protein acidic and rich in cysteine and inflammation: Another homeostatic property? Cytokine 2020, 133, 155179. [Google Scholar] [CrossRef] [PubMed]
- Ghanemi, A.; Yoshioka, M.; St-Amand, J. Secreted protein acidic and rich in cysteine and cancer: A homeostatic hormone? Cytokine 2020, 127, 154996. [Google Scholar] [CrossRef]
- Hood, D.A. Mechanisms of exercise-induced mitochondrial biogenesis in skeletal muscle. Appl. Physiol. Nutr. Metab. 2009, 34, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, N.J.; Parker, B.L.; Chaudhuri, R.; Fisher-Wellman, K.H.; Kleinert, M.; Humphrey, S.J.; Yang, P.; Holliday, M.; Trefely, S.; Fazakerley, D.J.; et al. Global Phosphoproteomic Analysis of Human Skeletal Muscle Reveals a Network of Exercise-Regulated Kinases and AMPK Substrates. Cell Metab. 2015, 22, 922–935. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, M.T.; Booth, F.W. Skeletal muscle adaptation to exercise: A century of progress. J. Appl. Physiol. (1985) 2000, 88, 327–331. [Google Scholar] [CrossRef]
- Melov, S.; Tarnopolsky, M.A.; Beckman, K.; Felkey, K.; Hubbard, A. Resistance exercise reverses aging in human skeletal muscle. PLoS ONE 2007, 2, e465. [Google Scholar] [CrossRef] [Green Version]
- Petriz, B.A.; Gomes, C.P.; Almeida, J.A.; de Oliveira, G.P., Jr.; Ribeiro, F.M.; Pereira, R.W.; Franco, O.L. The Effects of Acute and Chronic Exercise on Skeletal Muscle Proteome. J. Cell Physiol. 2017, 232, 257–269. [Google Scholar] [CrossRef]
- Holloway, K.V.; O′Gorman, M.; Woods, P.; Morton, J.P.; Evans, L.; Cable, N.T.; Goldspink, D.F.; Burniston, J.G. Proteomic investigation of changes in human vastus lateralis muscle in response to interval-Exercise training. Proteomics 2009, 9, 5155–5174. [Google Scholar] [CrossRef]
- Burniston, J.G. Changes in the rat skeletal muscle proteome induced by moderate-intensity endurance exercise. Biochim. Biophys. Acta 2008, 1784, 1077–1086. [Google Scholar] [CrossRef]
- Ruas, J.L.; White, J.P.; Rao, R.R.; Kleiner, S.; Brannan, K.T.; Harrison, B.C.; Greene, N.P.; Wu, J.; Estall, J.L.; Irving, B.A.; et al. A PGC-1alpha isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell 2012, 151, 1319–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coffey, V.G.; Hawley, J.A. Concurrent exercise training: Do opposites distract? J. Physiol. 2017, 595, 2883–2896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raue, U.; Trappe, T.A.; Estrem, S.T.; Qian, H.R.; Helvering, L.M.; Smith, R.C.; Trappe, S. Transcriptome signature of resistance exercise adaptations: Mixed muscle and fiber type specific profiles in young and old adults. J. Appl. Physiol. (1985) 2012, 112, 1625–1636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- St-Amand, J.; Yoshioka, M.; Nishida, Y.; Tobina, T.; Shono, N.; Tanaka, H. Effects of mild-exercise training cessation in human skeletal muscle. Eur. J. Appl. Physiol. 2012, 112, 853–869. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, K.A.; Krauss, R.M.; Fattor, J.A.; Horning, M.A.; Friedlander, A.L.; Bauer, T.A.; Hagobian, T.A.; Wolfel, E.E.; Brooks, G.A. Endurance training has little effect on active muscle free fatty acid, lipoprotein cholesterol, or triglyceride net balances. Am. J. Physiol. Endocrinol. Metab. 2006, 291, E656–E665. [Google Scholar] [CrossRef] [PubMed]
- Shono, N.; Urata, H.; Saltin, B.; Mizuno, M.; Harada, T.; Shindo, M.; Tanaka, H. Effects of low intensity aerobic training on skeletal muscle capillary and blood lipoprotein profiles. J. Atheroscler. Thromb. 2002, 9, 78–85. [Google Scholar] [CrossRef] [Green Version]
- Laguna-Camacho, A. Influence on Adiposity and Atherogenic Lipaemia of Fatty Meals and Snacks in Daily Life. J. Lipids 2017, 2017, 1375342. [Google Scholar] [CrossRef]
- Ribaroff, G.A.; Wastnedge, E.; Drake, A.J.; Sharpe, R.M.; Chambers, T.J.G. Animal models of maternal high fat diet exposure and effects on metabolism in offspring: A meta-regression analysis. Obes. Rev. 2017, 18, 673–686. [Google Scholar] [CrossRef]
- Flatt, J.P. The difference in the storage capacities for carbohydrate and for fat, and its implications in the regulation of body weight. Ann. N. Y. Acad. Sci. 1987, 499, 104–123. [Google Scholar] [CrossRef]
- Maher, T.; Clegg, M.E. Dietary lipids with potential to affect satiety: Mechanisms and evidence. Crit. Rev. Food Sci. Nutr. 2018, 1–26. [Google Scholar] [CrossRef]
- Lissner, L.; Levitsky, D.A.; Strupp, B.J.; Kalkwarf, H.J.; Roe, D.A. Dietary fat and the regulation of energy intake in human subjects. Am. J. Clin. Nutr. 1987, 46, 886–892. [Google Scholar] [CrossRef] [PubMed]
- Flatt, J.P.; Ravussin, E.; Acheson, K.J.; Jequier, E. Effects of dietary fat on postprandial substrate oxidation and on carbohydrate and fat balances. J. Clin. Investig. 1985, 76, 1019–1024. [Google Scholar] [CrossRef] [PubMed]
- Schutz, Y.; Flatt, J.P.; Jequier, E. Failure of dietary fat intake to promote fat oxidation: A factor favoring the development of obesity. Am. J. Clin. Nutr. 1989, 50, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Keleher, M.R.; Zaidi, R.; Shah, S.; Oakley, M.E.; Pavlatos, C.; El Idrissi, S.; Xing, X.; Li, D.; Wang, T.; Cheverud, J.M. Maternal high-fat diet associated with altered gene expression, DNA methylation, and obesity risk in mouse offspring. PLoS ONE 2018, 13, e0192606. [Google Scholar] [CrossRef] [Green Version]
- Keleher, M.R.; Zaidi, R.; Hicks, L.; Shah, S.; Xing, X.; Li, D.; Wang, T.; Cheverud, J.M. A high-Fat diet alters genome-Wide DNA methylation and gene expression in SM/J mice. BMC Genom. 2018, 19, 888. [Google Scholar] [CrossRef] [Green Version]
- Suter, M.A.; Ma, J.; Vuguin, P.M.; Hartil, K.; Fiallo, A.; Harris, R.A.; Charron, M.J.; Aagaard, K.M. In utero exposure to a maternal high-Fat diet alters the epigenetic histone code in a murine model. Am. J. Obstet. Gynecol. 2014, 210, 463.E1–463.E11. [Google Scholar] [CrossRef] [Green Version]
- Wankhade, U.D.; Zhong, Y.; Kang, P.; Alfaro, M.; Chintapalli, S.V.; Thakali, K.M.; Shankar, K. Enhanced offspring predisposition to steatohepatitis with maternal high-Fat diet is associated with epigenetic and microbiome alterations. PLoS ONE 2017, 12, e0175675. [Google Scholar] [CrossRef] [Green Version]
- Glendining, K.A.; Fisher, L.C.; Jasoni, C.L. Maternal high fat diet alters offspring epigenetic regulators, amygdala glutamatergic profile and anxiety. Psychoneuroendocrinology 2018, 96, 132–141. [Google Scholar] [CrossRef]
- Smilowitz, J.T.; German, J.B.; Zivkovic, A.M. Food Intake and Obesity: The Case of Fat. In Fat Detection: Taste, Texture, and Post Ingestive Effects; Montmayeur, J.P., le Coutre, J., Eds.; CRC Press/Taylor & Francis: Boca Raton, FL, USA, 2010. [Google Scholar]
- Astrup, A. The role of dietary fat in the prevention and treatment of obesity. Efficacy and safety of low-Fat diets. Int. J. Obes. Relat. Metab. Disord. J. Int. Assoc. Study Obes. 2001, 25 (Suppl. 1), S46–S50. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Wang, Q.; Liu, A.; Lan, X.; Huang, Y.; Zhao, Z.; Jie, H.; Chen, J.; Zhao, Y. Physiological Implications of Orexins/Hypocretins on Energy Metabolism and Adipose Tissue Development. ACS Omega 2020, 5, 547–555. [Google Scholar] [CrossRef] [Green Version]
- Milbank, E.; López, M. Orexins/Hypocretins: Key Regulators of Energy Homeostasis. Front. Endocrinol. 2019, 10, 830. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, M.; Bolduc, C.; Raymond, V.; St-Amand, J. High-Fat meal-Induced changes in the duodenum mucosa transcriptome. Obes. (Silver Spring Md.) 2008, 16, 2302–2307. [Google Scholar] [CrossRef] [PubMed]
- Mucunguzi, O.; Melouane, A.; Ghanemi, A.; Yoshioka, M.; Boivin, A.; Calvo, E.L.; St-Amand, J. Identification of the principal transcriptional regulators for low-fat and high-fat meal responsive genes in small intestine. Nutr. Metab. 2017, 14, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglass, J.D.; Malik, N.; Chon, S.H.; Wells, K.; Zhou, Y.X.; Choi, A.S.; Joseph, L.B.; Storch, J. Intestinal mucosal triacylglycerol accumulation secondary to decreased lipid secretion in obese and high fat fed mice. Front. Physiol. 2012, 3, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smolders, L.; Mensink, R.P.; Boekschoten, M.V.; de Ridder, R.J.J.; Plat, J. The acute effects on duodenal gene expression in healthy men following consumption of a low-Fat meal enriched with theobromine or fat. Sci. Rep. 2018, 8, 1700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, C.R.; Osadchiy, V.; Kalani, A.; Mayer, E.A. The Brain-Gut-Microbiome Axis. Cell Mol. Gastroenterol. Hepatol. 2018, 6, 133–148. [Google Scholar] [CrossRef] [Green Version]
- De Giorgio, M.R.; Yoshioka, M.; Riedl, I.; Moreault, O.; Cherizol, R.G.; Shah, A.A.; Blin, N.; Richard, D.; St-Amand, J. Trefoil factor family member 2 (Tff2) KO mice are protected from high-Fat diet-Induced obesity. Obes. (Silver Spring) 2013, 21, 1389–1395. [Google Scholar] [CrossRef]
- Ghanemi, A.; Melouane, A.; Mucunguzi, O.; Yoshioka, M.; St-Amand, J. Energy and metabolic pathways in trefoil factor family member 2 (Tff2) KO mice beyond the protection from high-Fat diet-Induced obesity. Life Sci. 2018, 215, 190–197. [Google Scholar] [CrossRef]
- De Giorgio, M.R.; Yoshioka, M.; St-Amand, J. Feeding regulates the expression of pancreatic genes in gastric mucosa. J. Obes. 2010, 2010, 371950. [Google Scholar] [CrossRef]
- Bolduc, C.; Yoshioka, M.; St-Amand, J. Acute molecular mechanisms responsive to feeding and meal constitution in mesenteric adipose tissue. Obes. (Silver Spring) 2010, 18, 410–413. [Google Scholar] [CrossRef]
- De Giorgio, M.R.; Yoshioka, M.; St-Amand, J. Feeding induced changes in the hypothalamic transcriptome. Clin. Chim. Acta 2009, 406, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Gondret, F.; Vincent, A.; Houee-Bigot, M.; Siegel, A.; Lagarrigue, S.; Louveau, I.; Causeur, D. Molecular alterations induced by a high-Fat high-Fiber diet in porcine adipose tissues: Variations according to the anatomical fat location. BMC Genom. 2016, 17, 120. [Google Scholar] [CrossRef] [Green Version]
- Moraes, R.C.; Blondet, A.; Birkenkamp-Demtroeder, K.; Tirard, J.; Orntoft, T.F.; Gertler, A.; Durand, P.; Naville, D.; Bégeot, M. Study of the Alteration of Gene Expression in Adipose Tissue of Diet-Induced Obese Mice by Microarray and Reverse Transcription-Polymerase Chain Reaction Analyses. Endocrinology 2003, 144, 4773–4782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Schindler, J.; Kanhere, A.; Edwards, L.; Allwood, J.W.; Dunn, W.B.; Schenk, S.; Philp, A. Exercise and high-fat feeding remodel transcript-metabolite interactive networks in mouse skeletal muscle. Sci. Rep. 2017, 7, 13485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hui, X.; Zhu, W.; Wang, Y.; Lam, K.S.; Zhang, J.; Wu, D.; Kraegen, E.W.; Li, Y.; Xu, A. Major urinary protein-1 increases energy expenditure and improves glucose intolerance through enhancing mitochondrial function in skeletal muscle of diabetic mice. J. Biol. Chem. 2009, 284, 14050–14057. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Rui, L. Major urinary protein regulation of chemical communication and nutrient metabolism. Vitam. Horm. 2010, 83, 151–163. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Jiang, L.; Rui, L. Identification of MUP1 as a regulator for glucose and lipid metabolism in mice. J. Biol. Chem. 2009, 284, 11152–11159. [Google Scholar] [CrossRef] [Green Version]
- van Schothorst, E.M.; Keijer, J.; Pennings, J.L.; Opperhuizen, A.; van den Brom, C.E.; Kohl, T.; Franssen-van Hal, N.L.; Hoebee, B. Adipose gene expression response of lean and obese mice to short-Term dietary restriction. Obes. (Silver Spring) 2006, 14, 974–979. [Google Scholar] [CrossRef] [Green Version]
- Harrell, C.S.; Burgado, J.; Kelly, S.D.; Johnson, Z.P.; Neigh, G.N. High-Fructose diet during periadolescent development increases depressive-Like behavior and remodels the hypothalamic transcriptome in male rats. Psychoneuroendocrinology 2015, 62, 252–264. [Google Scholar] [CrossRef] [Green Version]
- Ziotopoulou, M.; Mantzoros, C.S.; Hileman, S.M.; Flier, J.S. Differential expression of hypothalamic neuropeptides in the early phase of diet-Induced obesity in mice. Am. J. Physiol. Endocrinol. Metab. 2000, 279, E838–E845. [Google Scholar] [CrossRef] [Green Version]
- St-Amand, J.; Yoshioka, M.; Tanaka, K.; Nishida, Y. Transcriptome-Wide identification of preferentially expressed genes in the hypothalamus and pituitary gland. Front. Endocrinol. 2011, 2, 111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedard, K.; Bedard, J.; Rocheleau, G.; Ferland, G.; Gaudreau, P. Aging and diets regulate the rat anterior pituitary and hypothalamic transcriptome. Neuroendocrinology 2013, 97, 146–159. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, J.; Waki, S.; Matsumoto, R.; Odake, J.; Miyaji, T.; Tottori, J.; Iwanaga, T.; Iwahashi, H. Oligonucleotide microarray analysis of dietary-induced hyperlipidemia gene expression profiles in miniature pigs. PLoS ONE 2012, 7, e37581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maruthur, N.M.; Tseng, E.; Hutfless, S.; Wilson, L.M.; Suarez-Cuervo, C.; Berger, Z.; Chu, Y.; Iyoha, E.; Segal, J.B.; Bolen, S. Diabetes Medications as Monotherapy or Metformin-Based Combination Therapy for Type 2 Diabetes: A Systematic Review and Meta-analysis. Ann. Intern. Med. 2016, 164, 740–751. [Google Scholar] [CrossRef]
- Salomaki, H.; Heinaniemi, M.; Vahatalo, L.H.; Ailanen, L.; Eerola, K.; Ruohonen, S.T.; Pesonen, U.; Koulu, M. Prenatal metformin exposure in a maternal high fat diet mouse model alters the transcriptome and modifies the metabolic responses of the offspring. PLoS ONE 2014, 9, e115778. [Google Scholar] [CrossRef] [Green Version]
- Yoon, G.; Cho, K.A.; Song, J.; Kim, Y.-K. Transcriptomic Analysis of High Fat Diet Fed Mouse Brain Cortex. Front. Genet. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Hansen, D.; Dendale, P.; Berger, J.; van Loon, L.J.; Meeusen, R. The effects of exercise training on fat-mass loss in obese patients during energy intake restriction. Sports Med. 2007, 37, 31–46. [Google Scholar] [CrossRef]
- Miller, C.T.; Fraser, S.F.; Levinger, I.; Straznicky, N.E.; Dixon, J.B.; Reynolds, J.; Selig, S.E. The effects of exercise training in addition to energy restriction on functional capacities and body composition in obese adults during weight losss: A systematic review. PLoS ONE 2013, 8, e81692. [Google Scholar] [CrossRef] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghanemi, A.; Melouane, A.; Yoshioka, M.; St-Amand, J. Exercise and High-Fat Diet in Obesity: Functional Genomics Perspectives of Two Energy Homeostasis Pillars. Genes 2020, 11, 875. https://doi.org/10.3390/genes11080875
Ghanemi A, Melouane A, Yoshioka M, St-Amand J. Exercise and High-Fat Diet in Obesity: Functional Genomics Perspectives of Two Energy Homeostasis Pillars. Genes. 2020; 11(8):875. https://doi.org/10.3390/genes11080875
Chicago/Turabian StyleGhanemi, Abdelaziz, Aicha Melouane, Mayumi Yoshioka, and Jonny St-Amand. 2020. "Exercise and High-Fat Diet in Obesity: Functional Genomics Perspectives of Two Energy Homeostasis Pillars" Genes 11, no. 8: 875. https://doi.org/10.3390/genes11080875