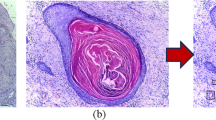
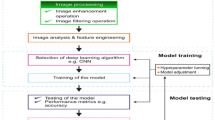
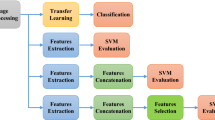
Abstract
The use of a binary classifier like the sigmoid function and loss functions reduces the accuracy of deep learning algorithms. This research aims to increase the accuracy of detecting and classifying oral tumours within a reduced processing time. The proposed system consists of a Convolutional neural network with a modified loss function to minimise the error in predicting and classifying oral tumours by reducing the overfitting of the data and supporting multi-class classification. The proposed solution was tested on data samples from multiple datasets with four kinds of oral tumours. The averages of the different accuracy values and processing times were calculated to derive the overall accuracy. Based on the obtained results, the proposed solution achieved an overall accuracy of 96.5%, which was almost 2.0% higher than the state-of-the-art solution with 94.5% accuracy. Similarly, the processing time has been reduced by 30–40 milliseconds against the state-of-the-art solution. The proposed system is focused on detecting oral tumours in the given magnetic resonance imaging (MRI) scan and classifying whether the tumours are benign or malignant. This study solves the issue of over fitting data during the training of neural networks and provides a method for multi-class classification.









Similar content being viewed by others
References
Al-Ma’aitah M, Ali AlZubi A (2018) Enhanced Computational Model for Gravitational Search Optimized Echo State Neural Networks Based Oral Cancer Detection. J Med Syst 42(11):205
Alsmadi MK (2018) A hybrid fuzzy C-means and Neutrosophic for jaw lesions segmentation. Ain Shams Eng J 9(4):697–706
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11(10):106
Anter AM, Ella HA (2018) CT liver tumor segmentation hybrid approach using neutrosophic sets, fast fuzzy c-means and adaptive watershed algorithm. Artif Intell Med 15(1):157
Aubreville M, Knipfer C, Oetter N, Jaremenko C, Rodner E, Denzler J, Bohr C, Neumann H, Stelzle F, Maier A (2017) Automatic classification of cancerous tissue in Laserendomicroscopy images of the Oral cavity using deep learning. Sci Rep 7(1):11979–11979
Bur AM et al Machine learning to predict occult nodal metastasis in early oral squamous cell carcinoma. Oral Oncol 92:20–25
Bura AM et al (2019) Machine learning to predict occult nodal metastasis in early oral squamous cell carcinoma. Oral Oncol 92(1):20–25
Das DK, Chakraborty C, Sawaimoon S, Maiti AK, Chatterjee S (2015) Automated identification of keratinization and keratin pearl area from in situ oral histological images. Tissue Cell 47(4):349–358
Das DK, Bose S, Maiti AK, Mitra B, Mukherjee G, Dutta PK (2018) Automatic identification of clinically relevant regions from oral tissue histological images for oral squamous cell carcinoma diagnosis. Tissue Cell 53:111–119
De Silva RK, Siriwardena BSMS, Samaranayaka A, Abeyasinghe WAMUL, Tilakaratne WM (2018) A model to predict nodal metastasis in patients with oral squamous cell carcinoma," (in eng). PLoS One 13(8):e0201755–e0201755
de Souza Tolentino E, Centurion BS, Ferreira LHC, de Souza AP, Damante JH, Rubira-Bullen IRF (2011) Oral adverse effects of head and neck radiotherapy: literature review and suggestion of a clinical oral care guideline for irradiated patients. J Appl Oral Sci 19(5):448–454
Halicek M, Lu G, Little JV, Wang X, Patel M, Griffith CC, el-Deiry MW, Chen AY, Fei B (2017) Deep convolutional neural networks for classifying head and neck cancer using hyperspectral imaging. J Biomed Opt 22(6):60503–60503
Halicek M, Little JV, Wang X, Chen AY, Fei B (2019) Optical biopsy of head and neck cancer using hyperspectral imaging and convolutional neural networks. J Biomed Opt 24(03):1
Health and Biology Data (2018) SkyMind, Ed., ed
Jain DK, Dubey SB, Choubey RK, Sinhal A, Arjaria SK, Jain A, Wang H (2018) An approach for hyperspectral image classification by optimizing SVM using self organizing map. J Comput Sci 25(1):252–259
Jeyaraj P, Nadar ERS (2019) Computer-assisted medical image classification for early diagnosis of oral cancer employing deep learning algorithm. J Cancer Res Clin Oncol 145(4):1–9
Li H, Huang Y, Zhang Z (2017) An improved faster R-CNN for same object retrieval. IEEE 5:13665–13676
Li H et al (2018) An improved deep learning approach for detection of thyroid papillary cancer in ultrasound images. Sci Rep 8(1):6600
Liang S, Tang F, Huang X, Yang K, Zhong T, Hu R, Liu S, Yuan X, Zhang Y (2019) Deep-learning-based detection and segmentation of organs at risk in nasopharyngeal carcinoma computed tomographic images for radiotherapy planning. Eur Radiol 29(4):1961–1967
Lu G, Fei B (2014) Medical hyperspectral imaging: a review. J Biomed Opt 19(1):10901–10901
Mohammed MA, Ghani MKA, Hamed RI, Ibrahim DA (2017) Review on nasopharyngeal carcinoma: concepts, methods of analysis, segmentation, classification, prediction and impact: a review of the research literature. J Comput Sci 21(1):283–298
Obermeyer Z, Emanuel EJ (2016) Predicting the future — big data, machine learning, and clinical medicine. N Engl J Med 375:1216–1219
Oetter N et al (2016) Development and validation of a classification and scoring system for the diagnosis of oral squamous cell carcinomas through confocal laser endomicroscopy. J Transl Med 14(1):159
Poedjiastoeti W, Suebnukarn S (2018) Application of convolutional neural network in the diagnosis of jaw tumors. HealthCare Informat Res 24(3):236–241
Sharma N, Om H (2014) Extracting significant patterns for oral cancer detection using apriori algorithm. Intell Inf Manag 6(2):30–37
Shi M, Zhang B (2011) Semi-supervised learning improves gene expression-based prediction of cancer recurrence. Bioinformatics 27(21):3017–3023
Shin H-C, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM (2016) Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 35(5):1285–1298
Tong L-I, Chang Y-C, Lin S-H (2011) Determining the optimal re-sampling strategy for a classification model with imbalanced data using design of experiments and response surface methodologies. Expert Syst Appl 38(4):4222–4227
Xiao Y, Wu J, Lin Z, Zhao X (2018) A deep learning-based multi-model ensemble method for cancer prediction. Comput Methods Prog Biomed 153(C):1–9
Xiao Y, Wu J, Lin Z, Zhao X (2018) A semi-supervised deep learning method based on stacked sparse auto-encoder for cancer prediction using RNA-seq data. Comput Methods Prog Biomed 166(1):99–105
Yuan X, Xie L, Abouelenien M (2018) A regularized ensemble framework of deep learning for cancer detection from multi-class, imbalanced training data. Pattern Recogn 77(1):160–172
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bhandari, B., Alsadoon, A., Prasad, P.W.C. et al. Deep learning neural network for texture feature extraction in oral cancer: enhanced loss function. Multimed Tools Appl 79, 27867–27890 (2020). https://doi.org/10.1007/s11042-020-09384-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-020-09384-6