Abstract
Objectives
To develop and evaluate an automated method for prostate T2-weighted (T2W) image normalization using dual-reference (fat and muscle) tissue.
Materials and methods
Transverse T2W images from the publicly available PROMISE12 (N = 80) and PROSTATEx (N = 202) challenge datasets, and an in-house collected dataset (N = 60) were used. Aggregate channel features object detectors were trained to detect reference fat and muscle tissue regions, which were processed and utilized to normalize the 3D images by linear scaling. Mean prostate pseudo T2 values after normalization were compared to literature values. Inter-patient histogram intersections of voxel intensities in the prostate were compared between our approach, the original images, and other commonly used normalization methods. Healthy vs. malignant tissue classification performance was compared before and after normalization.
Results
The prostate pseudo T2 values of the three tested datasets (mean ± standard deviation = 78.49 ± 9.42, 79.69 ± 6.34 and 79.29 ± 6.30 ms) corresponded well to T2 values from literature (80 ± 34 ms). Our normalization approach resulted in significantly higher (p < 0.001) inter-patient histogram intersections (median = 0.746) than the original images (median = 0.417) and most other normalization methods. Healthy vs. malignant classification also improved significantly (p < 0.001) in peripheral (AUC 0.826 vs. 0.769) and transition (AUC 0.743 vs. 0.678) zones.
Conclusion
An automated dual-reference tissue normalization of T2W images could help improve the quantitative assessment of prostate cancer.
Similar content being viewed by others
Introduction
Prostate cancer is the second most commonly diagnosed cancer and the leading cause of cancer-related deaths among men worldwide [1]. Multiparametric magnetic resonance imaging (mpMRI) has been established as a valuable diagnostic tool for prostate cancer [2, 3]. T2-weighted (T2W) MR imaging is considered an essential pillar of mpMRI for prostate cancer diagnosis due to the high spatial resolution and the superior anatomical details it provides [3,4,5]. However, unlike other mpMRI sequences such as diffusion-weighted and dynamic contrast-enhanced imaging, the use of T2W imaging has mainly been limited to a qualitative evaluation of prostate anomalies. Its utility for quantitative analysis is hindered by, among other things, non-standard signal intensity (SI) attributed to scanner parameters such as the field strength, coil type, signal amplification, and acquisition protocols [6,7,8,9]. To make use of T2W images for quantitative analysis, an image processing step called SI normalization is often required, which theoretically removes the variation in SI between images from different scan sessions. Consequently, SI normalization enables comparing T2W image values from different patients (inter-patient comparison), patient follow-up at multiple scans over time (intra-patient comparison), and tissue classification tasks in the setting of a radiomics or computer-assisted diagnosis approach [10, 11].
SI normalization is not new, and over the years, different approaches have been proposed for prostate imaging. Due to their simplicity, histogram-based approaches, which typically depend on pre-set histogram landmarks to deform or rescale the SI [7, 12], have become the most commonly used [10, 13,14,15,16]. A drawback of these methods is that they usually rely on the content in the complete 2D or 3D image, which is subject to variation due to differences in scan settings (e.g. the field-of-view) and patient-related factors (e.g. bladder filling). Recently, SI normalization utilizing single or multiple reference tissues has shown promise as an alternative to histogram-based methods [17,18,19,20,21]. In single reference tissue normalization, the original T2W image SI is scaled by the SI in the corresponding reference tissue region-of-interest (ROI). One common example of this in the prostate is normalization to the SI of the obturator internus or levator ani muscles [17, 22,23,24]. Multi-reference tissue normalization, on the other hand, utilizes the SIs of multiple reference tissues to create a linear or non-linear regression model to estimate the normalized T2W image values [18, 19]. The assumption is that reference tissue-based normalization is less sensitive to variations in scan settings and patient-related factors. However, a key aspect of this approach is labelling the reference tissues, to enable SI extraction. Currently, this is done manually, which is a time-consuming and tedious process. Automated delineation of reference tissue ROIs would make the approach more efficient and could possibly facilitate its integration into clinical practice. This can for example be achieved using automated semantic segmentation or object detection methods. In comparison with semantic segmentation, object detection requires less processing power, time and data [25, 26].
The contribution of this work is a novel method for automated dual-reference tissue normalization of T2W images of the prostate, based on object recognition to extract the reference tissue ROIs. We compared the automatically extracted reference tissue intensities with those of manually delineated ROIs, and evaluated the merit of the proposed method for inter- and intra-patient comparison of T2W image intensities and for the classification of malignant lesions versus healthy prostate tissue.
Materials and methods
Datasets
In this study, transverse T2W images from three separate datasets were used: the PROMISE12 grand challenge dataset (N = 80) [27], the PROSTATEx challenge dataset (N = 202) [28] and a dataset of in-house collected T2W images from patients who underwent two sequential MRI scans for detection and biopsy-guiding, respectively (N = 60). The Regional Committee for Medical and Health Research Ethics (REC Mid Norway) approved the use of the in-house collected dataset (identifier 2017/576) and granted permission for passive consent to be used, whereas the two other datasets were publicly available.
The PROMISE12 dataset [27] consists of multi-centre and multi-vendor transverse T2W images obtained with different field strengths, acquisition protocols and coils. It also includes manual expert segmentations of the whole prostate for 50 cases. The PROSTATEx challenge dataset [28] consists of pre-biopsy mpMRI sequences acquired at Radboud University Medical Centre, Nijmegen, Netherlands. The whole prostate, peripheral zone, and cancer-suspicious volumes of interest (VOIs) were manually delineated by radiologists (at Miller School of Medicine, Miami, FL, USA) based on targeted biopsy locations provided by the challenge organizers. The presence of clinically significant prostate cancer (Gleason score > 3 + 3) in the targeted biopsy cores was then used to label each cancer-suspicious VOI as a true positive (malignant) or false positive radiological finding. The rest of the prostate was considered healthy tissue.
The in-house collected dataset was obtained from St. Olavs Hospital, Trondheim University Hospital, Trondheim, Norway between March 2015 and December 2017. It consists of pairs of pre-biopsy 3 T images from 60 patients (median age = 65.5 years; range 47–75 years) acquired at two different time points: first, at the initial visit for detection of prostate cancer (scan 1), and second, during an MR-guided biopsy procedure (scan 2). The interval between scans ranged 1–71 days with a median interval of 7 days. T2W imaging was performed on a Magnetom Skyra 3 T MRI system (Siemens, Erlangen, Germany) with a turbo spin-echo sequence (Scan 1: repetition time/echo time = 4800–9520/104 ms, 320 × 320 – 384 × 384 matrix size, 26–32 slices, 3 mm slice thickness and 0.5 × 0.5–0.6 × 0.6 mm2 in plane resolution. Scan 2: repetition time/echo time = 5660–7740/101–104 ms, 320 × 320–384 × 384 matrix size, 19–26 slices, 3 mm slice thickness and 0.5 × 0.5–0.6 × 0.6 mm2 in plane resolution). The whole prostate volumes were manually delineated by a radiologist in training.
Proposed intensity normalization method
Figure 1 gives an overview of the proposed method, termed AutoRef. The method contains several tuneable parameters, which were optimized as described in the next section. In the final, optimized version, the 3D T2W images were first pre-processed, which included N4 bias field correction [29], rescaling to the 99th percentile intensity value and resizing the transverse slices to 384 × 384 pixels with 0.5 × 0.5 mm in-plane resolution. Two separate aggregate channel features (ACF) object detectors [25] were then trained, using two training stages for the iterative training process, to detect rectangular ROIs containing fat and muscle (levator ani muscle) tissue on the 2D transverse slices. Both object detectors were forced to focus on regions where the ROIs were expected to minimize the detection of unwanted structures. For fat, the focus region comprised the lower (posterior) 50% of the image in the lower (inferior) 75% of the slices. For muscle, the focus region comprised the middle (posterior-anterior) 50% of the image in the middle (inferior-superior) 50% of the slices. The three slices containing the rectangular ROIs with the highest probability of fat/muscle were identified, and post-processed by Otsu thresholding [30] and morphological opening, with disk shape of one-pixel radius, to extract the largest connected bright and dark structures in the detected rectangle, representing fat and muscle ROIs, respectively. The fat (\(I^{{{\text{fat}}}}\)) and muscle (\(I^{{{\text{muscle}}}} )\) reference intensity values were then calculated as the 90th and 10th percentiles, respectively, of the intensity values in these ROIs. Subsequently, the 3D image intensities (\(I\left( {x, y, z} \right)\)) were normalized to pseudo T2 values (\(pT2\left( {x, y, z} \right)\)) by linearly scaling \(I^{{{\text{fat}}}}\) and \(I^{{{\text{muscle}}}}\) to their respective T2 values at 3 T from literature (\(T2^{{{\text{fat}}}}\) = 121 ms and \(T2^{{{\text{muscle}}}}\) = 40 ms) [31], using Eq. (1):
Overview of AutoRef, the proposed normalization method. The T2W images were first pre-processed including bias field correction, rescaling and resizing. Rectangles containing fat/muscle were then detected slice by slice using trained aggregate channel features (ACF) detectors. The three slices containing rectangular regions with the highest probability of containing fat/muscle were identified and post-processed by Otsu thresholding and morphological opening to extract the largest connected fat/muscle region-of-interest (ROI). From these ROIs, fat/muscle reference intensities were obtained for normalization of the 3D image intensities
Training, validation and testing
The PROMISE12 dataset was shuffled and split for training (N = 40), validation (N = 20), and testing (N = 20) of AutoRef. Since prostate segmentations were only available for 50 cases, the splitting was semi-random and controlled in a way that ensured that only cases with the required segmentations were included in the validation and test subsets. The PROSTATEx and the in-house collected datasets were used for testing only.
The training and validation subsets were used to train the object detectors and to find the optimal pre- and post-processing settings resulting in the best performance of AutoRef. An overview of the optimization results in the validation subset is provided in Online Resource 1. The trained detectors and optimal parameter settings, as described in the previous section, were subsequently applied to normalize the images in the PROMISE12 test subset, the PROSTATEx dataset and the in-house collected dataset.
Verification of reference tissue intensities
The reference tissue intensities extracted from muscle and fat tissue by AutoRef, \(I^{{{\text{fat}}}}\) and \(I^{{{\text{muscle}}}}\), respectively, were compared with those of manually drawn ROIs in the PROMISE12 test subset. In the manual approach, a researcher with 3 years of experience with prostate imaging (MRSS) delineated three ROIs in both fat and muscle tissue on what were judged to be representative T2W slices by visual inspection. The 90th and 10th percentiles of the intensity values within the manual fat and muscle ROIs, respectively, were compared to \(I^{{{\text{fat}}}}\) and \(I^{{{\text{muscle}}}}\) and the relative differences and absolute relative differences were calculated. Visual inspection of all automatically extracted fat and muscle ROIs from the PROMISE12 test subset, the PROSTATEx dataset, and the in-house collected dataset was performed by the same researcher to reveal any suboptimal ROIs. A ROI was considered suboptimal when it failed to detect the tissue of interest or covered additional regions not belonging to fat or muscle on any of the three slices.
Inter- and intra-patient performance of normalization
The performance of AutoRef was compared to the original images and three other automated normalization methods, commonly used in literature, i.e. histogram stretching (Eq. (2)) [8], histogram equalization (histeq function from MATLAB®), and Gaussian kernel normalization (Eq. (3)) [8]:
where \(I_{\max }\) and \(I_{\min }\) represent the maximum and minimum intensity values, respectively, in the original image \(I\).
where \(\mu\) and \(\sigma\) represent the mean and standard deviation of the voxel intensities in the original image \(I\), respectively.
Furthermore, the performance of AutoRef using two reference tissues (as proposed) was compared to that of AutoRefmuscle (Eq. 4), which uses only muscle reference intensity values, as by several other studies [17, 22,23,24]:
where \(I^{{{\text{muscle}}}}\) represents the mean value of the automatically extracted muscle ROIs and \(T2^{{{\text{muscle}}}}\) the muscle T2 value from literature.
The histogram intersections (Eq. 5) of whole prostate voxel intensities of each pair of patients within the PROMISE12 test subset were used as a metric of inter-patient performance. In addition, the PROSTATEx dataset was used to separately evaluate the inter-patient histogram intersections in the peripheral (PZ) and transition zone (TZ):
where \(H_{x}\) and \(H_{y}\) represent the intensity histograms of patient \(x\) and patient \(y\), respectively, and \(n\) represents the number of histogram bins (set to 100). \(H_{x}\) and \(H_{y}\) were normalized to the total number of voxels in the prostate or zone.
The in-house collected dataset was used to assess the intra-patient performance, by measuring the whole prostate histogram intersection between the pair of consecutive scans of the same patient (Eq. 6):
where \(H_{1}\) and \(H_{2}\) represent the histograms for the first and second scans of the same patient, respectively, and \(n\) represents the number of histogram bins (set to 100). \(H_{1}\) and \(H_{1}\) were normalized to the total number of voxels in the prostate.
For all datasets, the \(pT2\left( {x, y, z} \right)\) values of prostate tissue obtained with AutoRef and AutoRefmuscle were compared to T2 values from the literature [31]. Furthermore, the \(pT2\left( {x, y, z} \right)\) values of prostate tissue obtained with AutoRef were compared between patients scanned with and without an endorectal coil.
Classification of malignant lesions versus healthy prostate tissue
Mean intensity values were extracted from the histologically verified malignant lesions and from healthy tissue in the PZ and TZ of the PROSTATEx dataset. The values were used as predictors in logistic regression models to distinguish healthy prostate tissue from malignant lesions in the PZ and TZ, separately. To ensure representative results least influenced by how the data was split, the models were trained and tested using 10 iterations with fivefold cross-validation. In each iteration, the dataset was randomly split, in a controlled way, into training (4 folds) and testing (1 fold) datasets, allowing each fold to be used once for testing. Receiver operating characteristic (ROC) curves were created to evaluate the performance of the classifier at each iteration and the mean and 95% confidence interval (CI) of the area under the curves (AUC) was reported.
Statistical analysis
Wilcoxon signed-rank tests were used to assess statistical differences between the manually and automatically obtained reference tissue intensities, and between the histogram intersections of the various normalization methods. Two-sample t tests were used to assess statistical differences between the pseudo T2 and literature T2 values of the prostate [31], and between the prostate pseudo T2 values of patients scanned with and without an endorectal coil. Wilcoxon rank-sum tests were used to assess statistical differences between the mean intensity values of healthy and malignant regions after normalization. DeLong’s method [32] was used to assess statistical differences between AUCs. The tests were followed by Benjamini–Hochberg correction for multiple comparisons [33] with false discovery rate of 0.05. Corrected p values less than 0.05 were considered statistically significant.
All algorithms and analyses were implemented and performed in MATLAB R2019b (The Mathworks, Nattick, MA, USA). The proposed algorithm will be made available on GitHub at https://github.com/ntnu-mr-cancer/AutoRef.
Results
Verification of reference tissue intensities
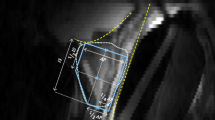
Figure 2a shows the manually and automatically extracted fat and muscle intensities, respectively, for all cases in the PROMISE12 test subset before normalization. There were significant differences between the reference intensity values from manually and automatically detected fat (p = 0.048) and muscle (p = 0.018) ROIs, with relative differences (median (range)) of 2.52% (− 16.21 to 39.86%) for fat and 7.03% (− 20.24 to 23.20%) for muscle. The absolute relative differences [median (range)] between the manual and automated approach were 5.25% (0.17–39.86%) for fat and 9.10% (1.74–23.20%) for muscle intensities. Visual inspection revealed that automated ROIs were suboptimal in 4/20 (20%), 4/202 (2%) and 0/120 (0%) cases for fat and in 0/20 (0%), 3/202 (1.5%) and 0/120 (0%) for muscle ROIs in the PROMISE12 test subset, the PROSTATEx dataset and the in-house collected dataset, respectively, whereas the method performed well in all other cases. In the PROMISE12 test subset, 3/4 (75%) suboptimal ROIs were found in patients with an endorectal coil. Figure 2b shows representative examples of optimal ROIs automatically extracted with our method. All automatically extracted suboptimal ROIs are shown in Online Resource 2. It can be appreciated that the ‘suboptimal parts’ of the ROIs are often relatively small and of similar image intensity compared to the ‘correct parts’ of the ROIs, so their impact on the normalization is limited as shown in Online Resource 2.
Inter- and intra-patient evaluation of normalization performance
Figure 3 shows examples from the PROMISE12 test subset, the PROSTATEx dataset, and the in-house collected dataset before and after normalization using AutoRef. The image intensities are more homogeneous within and between the datasets after normalization. This improvement is most obvious in the PROMISE12 dataset, which was acquired with varying protocols, field strengths, and at multiple centres.
Central slice through the prostates of five patients from the PROMISE12 test subset, the PROSTATEx dataset and the in-house collected dataset before (left panel) and after normalization (right panel). In both panels, the images were window-levelled from 0 to 2 times the mean prostate intensity of all images in the respective dataset
The intensity histograms from the original and normalized images of PROMISE12 test subset are displayed in Online Resource 3. Figure 4a and Table 1 show that AutoRef resulted in significantly higher inter-patient intersections than the original data and the other normalization methods, except for AutoRefmuscle.
The inter-patient histogram intersections of the proposed method (AutoRef) compared to original and normalized images for the whole prostate (a), the peripheral (PZ; b) and transitional zone (TZ; c), respectively. The PROMISE12 test subset and PROSTATEx dataset were used in a, and b and c, respectively. AutoRef intersections were significantly higher (p < 0.001) than others, except for AutoRefmuscle in a (p = 0.424) and histogram stretching in c (p = 0.154). The histogram intersections between scan 1 and scan 2 of the in-house collected dataset (d) of AutoRef were significantly higher than for histogram equalization (p < 0.001), but similar to those of the other methods
Figure 4b, c and Table 1 also present the inter-patient histogram intersections for PZ and TZ of the PROSTATEx dataset. In both zones, the histogram intersections after normalization with AutoRef were significantly higher than those of the original data and the other normalization methods, except for histogram stretching in TZ.
The intra-patient histogram intersections between scan 1 and scan 2 of the in-house collected dataset are shown in Fig. 4c and Table 2. AutoRef resulted in significantly higher intra-patient intersections than histogram equalization but performed similar to the original data and the other normalization methods.
Figure 5 compares the pseudo T2 values of the whole prostate obtained with AutoRef and AutoRefmuscle with those reported in the literature (80 ± 34 ms) [31]. Using AutoRef, the mean ± standard deviation prostate pseudo T2 values were 78.49 ± 9.42 ms (p = 0.063), 79.69 ± 6.34 ms (p = 0.486) and 79.29 ± 6.30 ms (p = 0.161) for PROMISE12 test subset, the PROSTATEx dataset and the in-house collected dataset, respectively. Pseudo T2 values were not significantly different between patients scanned with (83.15 ± 8.85 ms) or without (79.36 ± 6.41 ms) an endorectal coil (p = 0.690). Using AutoRefmuscle, the prostate pseudo T2 values were significantly higher (p < 0.001) than literature values for all the datasets.
Box and whisker plots of the mean prostate pseudo T2 values of the patients in the PROMISE12 test subset, the PROSTATEx dataset and the in-house collected dataset after normalization with the proposed dual-reference normalization method (AutoRef) and single reference tissue normalization (AutoRefmuscle). The dashed lines correspond to the T2 values reported in literature. All the mean prostate T2 values for AutoRefmuscle, but not AutoRef, were significantly higher than those reported in literature (p < 0.001)
Classification of malignant lesions versus healthy prostate tissue
Figure 6a, b and Table 3 compare the performances (ROC curves and mean AUCs of the 10 iterations, respectively) of AutoRef and other methods in the classification of healthy tissue versus biopsy-confirmed cancer regions. In the PZ, AutoRef performed significantly better than the original data and the other normalization methods. In the TZ, the performance was similar to Gaussian kernel normalization and AutoRefmuscle, but significantly better than the original data, histogram stretching and histogram equalization. Figure 6c shows box and whisker plots of the mean pseudo T2 values of healthy and malignant regions after AutoRef normalization, which were significantly different in both the PZ and TZ (p < 0.001).
The receiver operating characteristic curves and areas under the curves (AUC; mean of 10 iterations) for the proposed method (AutoRef), the original images and the other investigated normalization methods in the peripheral (PZ; a) and transitional zone (TZ; b). In PZ, the AUC for AutoRef was significantly higher than that of the other methods (p < 0.001), whereas in TZ it was significantly higher than the original data (p < 0.001), histogram stretching (p = 0.010) and histogram equalization (p = 0.007). The mean pseudo T2 values (c) were significantly different between healthy and malignant regions in both the PZ and TZ. (p < 0.001)
Discussion
In this paper, we propose a new method for automated dual-reference tissue normalization of T2W images of the prostate, which shows promise for quantitative assessment of prostate cancer and could ease the comparison of T2-weighted images between and within patients. The proposed method successfully uses a simple object detector to extract reference tissue intensities from fat and muscle surrounding the prostate, which are subsequently used for intensity normalization of the 3D T2-weighted image. The proposed method generally resulted in higher inter-patient histogram intersections compared to the other investigated automated normalization methods, which indicates that the normalized intensity values in the prostate are more similar between images. Furthermore, the proposed method resulted in images with pseudo T2 values comparable to T2 values reported in the literature [31]. Lastly, as demonstrated by the improved classification of healthy versus malignant tissue, the proposed method successfully reduced the inter-patient variation in T2W image intensities, which could facilitate the extraction and application of meaningful intensity-based image features for quantitative assessment of prostate cancer, e.g. in a radiomics or computer-assisted diagnosis framework [34].
T2W normalization is paramount for the quantitative assessment of prostate cancer, and several methods have been previously proposed in the literature. Liu et al. [13] defined a non-parametric normalization standard as the median image intensity plus two times the inter-quartile range. Artan et al. [14] and Ozer et al. [15] normalized T2W images in a way similar to the Gaussian kernel method investigated here, but with the mean and standard deviation extracted from the PZ instead of the entire image. However, these methods require manual delineation of the PZ and might not be valid if the image intensities do not follow a Gaussian distribution [10]. Lemaitre et al. [10] chose to normalize the images using a parametric model assuming a Rician distribution of the voxel intensities in the whole prostate. Yet differently, Nyúl et al. [7] proposed a two-stage method, wherein the first stage a template histogram with landmarks of interest is created and in the second stage new histograms are mapped via linear transformation to the template. This method assumes that the MR images of the same sequence should have the same intensity distribution, which might not be the case for varying protocols. Vos et al. proposed a sequence-based approach, which depends on the original T2W signal, proton density value, a reference tissue, and a known sequence model to estimate new normalized T2W images [35]. Although this approach performs well, the intricate nature and additional scan time make its practical implementation difficult. Niaf et al. [20, 21] investigated a single reference tissue method that normalizes the image intensities by dividing by the mean intensity value of the bladder. Likewise, Peng et al. [17] normalized the images separately using each of the levator ani muscle, urinary bladder, and pubic bone, and concluded that using levator ani muscle as a single reference tissue gave the best results. In this work, the performance of AutoRef using only muscle reference intensities was shown to be generally inferior to that based on a dual-reference tissue normalization approach, and unable to correctly map the image intensities to literature T2 values. Our method uses fat as a second reference tissue because it typically has high T2W intensity values, thus together with muscle covering the full range of expected prostate intensity values, it is present in all images and less vulnerable to external factors than for example the urinary bladder.
Recently, Stoilescu et al. [19] showed that multi-reference tissue normalization of T2W prostate images significantly improved prostate cancer classification accuracy in comparison to non-normalized images. Four reference tissues were used based on manually annotated ROIs, which currently hinders the implementation of the method in clinical practice. Therefore, in our work, we developed an automated approach for detecting ROIs to enable multi-reference tissue normalization using two reference tissues (fat and levator ani muscle). The ACF detector used in this work is a relatively simple, classical machine learning approach that was able to accurately detect the fat and muscle ROIs in nearly all cases, despite the small training dataset (N = 40). Exceptions were found in 8/342 (2%) cases for fat and 3/342 (1%) cases for muscle ROIs when considering all patients, and in 1/331 (0.3%) and 3/331 (1%) cases, respectively, when considering patients scanned without an endorectal coil. The detection of fat thus performs worse in patients scanned with an endorectal coil but this may not pose a problem in clinical practice, as 3 T MRI with body surface coils is currently the recommended and preferred method. The detection of both fat and muscle may be further improved by using a larger dataset for training, while the method can also be extended to include more reference tissues if deemed necessary, which is subject of further investigation.
Although AutoRef generally performed better than or similar to the other normalization methods in all datasets, the largest differences were observed in the multi-centre, multi-vendor PROMISE12 dataset. In this dataset, images were acquired with 1.5 T or 3 T scanners, with or without an endorectal coil, and with different acquisition protocols, all of which are likely to influence the T2W image intensity. An important advantage of our method to the other investigated normalization methods is that the image intensities could be correctly mapped to literature T2 values [31], irrespective of these factors. The pseudo T2 values could be an interesting alternative to quantitative T2 mapping, given the limited scan time available in clinical practice, but this needs further investigation in studies where T2 maps are also acquired. However, it should be noted that AutoRef does not correct for local differences in signal intensities caused by the non-uniform sensitivity of the receiver coils. This effect is especially apparent for images acquired with an endorectal coil, which typically shows an intensity profile inversely related to proximity to the coil. Although we showed that the mean pseudo T2 values of images acquired with an endorectal coil were comparable to those acquired with body surface coils, there may be differences in intensity distribution within the prostate gland that are not accounted for by AutoRef.
In the intra-patient evaluation, AutoRef had similar intra-patient histogram intersections compared to the original data and most of the other investigated methods. This probably reflects the limited variability in the in-house collected dataset, which has been acquired at the same centre, the same scanner, with the same protocols at a relatively short interval between scans. It would be insightful to assess the performance of the method in a dataset where the same patients are systematically scanned at different hospitals, but such data are probably scarce.
Normalization with the proposed method resulted in a significantly higher AUC for the classification of histologically verified PZ lesions compared to the other methods. For TZ lesions, the AUC was significantly higher than the original data, histogram stretching and histogram equalization, and on par with the other normalization methods. However, the differences in the classification performance were relatively small, which again may be the result of the limited variability in a dataset acquired at a single centre and with a single protocol [28]. Furthermore, considerable overlap in pseudo T2 values was still present between healthy tissue and malignant lesions, especially in the TZ, indicating that pseudo T2 values alone may not be sufficient to detect prostate cancer in clinical practice.
Our study has some limitations. Quantitative T2 maps were not available for the patients included in this study, which hindered a direct comparison of the pseudo T2 values with a gold standard. Although we included several commonly applied automated normalization methods in this study, there are still many more described in the literature, as discussed above, that may perform better than those included here. In addition, it would be interesting to compare the performance of the proposed object detector to that of semantic segmentation for detecting ROIs, which will be subject to further research. Despite these limitations, we have shown that our proposed method for automated dual-reference tissue normalization performed equal to or better than other automated normalization methods. The method requires no manual input and the resulting images can be used for both quantitative and qualitative assessment of prostate cancer.
Conclusion
We successfully developed a method for automated dual-reference tissue normalization of T2W MR images of the prostate using object recognition. The method was shown to reduce T2W intensity variation between scans and could improve the quantitative assessment of prostate cancer on MRI.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424
Barentsz JO, Richenberg J, Clements R, Choyke P, Verma S, Villeirs G, Rouviere O, Logager V, Futterer JJ (2012) ESUR prostate MR guidelines 2012. Eur Radiol 22(4):746–757
Weinreb JC, Barentsz JO, Choyke PL, Cornud F, Haider MA, Macura KJ, Margolis D, Schnall MD, Shtern F, Tempany CM, Thoeny HC, Verma S (2016) PI-RADS prostate imaging—reporting and data system: 2015, version 2. Eur Urol 69(1):16–40
Hoeks CM, Barentsz JO, Hambrock T, Yakar D, Somford DM, Heijmink SW, Scheenen TW, Vos PC, Huisman H, van Oort IM, Witjes JA, Heerschap A, Futterer JJ (2011) Prostate cancer: multiparametric MR imaging for detection, localization, and staging. Radiology 261(1):46–66
Wang S, Burtt K, Turkbey B, Choyke P, Summers RM (2014) Computer aided-diagnosis of prostate cancer on multiparametric MRI: a technical review of current research. Biomed Res Int 2014:789561
Simmons A, Tofts PS, Barker GJ, Arridge SR (1994) Sources of intensity nonuniformity in spin-echo images at 1.5-T. Magn Reson Med 32(1):121–128
Nyul LG, Udupa JK, Zhang X (2000) New variants of a method of MRI scale standardization. IEEE Trans Med Imaging 19(2):143–150
Loizou CP, Pantziaris M, Seimenis I, Pattichis CS (2009) Brain MR image normalization in texture analysis of multiple sclerosis. In: Proceedings of the 9th international conference on information technology and applications in biomedicine, Larnaca, p 131
Madabhushi A, Udupa JK (2006) New methods of MR image intensity standardization via generalized scale. Med Phys 33(9):3426–3434
Lemaitre G, Rastgoo M, Massich J, Vilanova JC, Walker PM, Freixenet J, Meyer-Baese A, Meriaudeau F, Marti R (2015) Normalization of T2W-MRI prostate images using Rician a priori. In: Proceedings of SPIE medical imaging, San Diego, p 978529
Schwier M, van Griethuysen J, Vangel MG, Pieper S, Peled S, Tempany C, Aerts H, Kikinis R, Fennessy FM, Fedorov A (2019) Repeatability of multiparametric prostate MRI radiomics features. Sci Rep 9(1):9441
Ge YL, Udupa JK, Nyul LG, Wei LG, Grossman RI (2000) Numerical tissue characterization in MS via standardization of the MR image intensity scale. J Magn Reson Imaging 12(5):715–721
Liu P, Wang SJ, Turkbey B, Grant K, Pinto P, Choyke P, Wood BJ, Summers RM (2013) A prostate cancer computer-aided diagnosis system using multimodal magnetic resonance imaging and targeted biopsy labels. In: Proceedings of SPIE medical imaging, Lake Buena Vista, p 86701G
Artan Y, Haider MA, Langer DL, van der Kwast TH, Evans AJ, Yang YY, Wernick MN, Trachtenberg J, Yetik IS (2010) Prostate cancer localization with multispectral MRI using cost-sensitive support vector machines and conditional random fields. IEEE Trans Image Process 19(9):2444–2455
Ozer S, Langer DL, Liu X, Haider MA, van der Kwast TH, Evans AJ, Yang YY, Wernick MN, Yetik IS (2010) Supervised and unsupervised methods for prostate cancer segmentation with multispectral MRI. Med Phys 37(4):1873–1883
Lv DJ, Guo XM, Wang XY, Zhang J, Fang J (2009) Computerized characterization of prostate cancer by fractal analysis in MR images. J Magn Reson Imaging 30(1):161–168
Peng YH, Jiang YL, Oto A (2014) Reference-tissue correction of T-2-weighted signal intensity for prostate cancer detection. In: Proceedings of SPIE medical imaging, San Diego, p 903508
Leung KK, Clarkson MJ, Bartlett JW, Clegg S, Jack CR, Weiner MW, Fox NC, Ourselin S, AsDN I (2010) Robust atrophy rate measurement in Alzheimer's disease using multi-site serial MRI: tissue-specific intensity normalization and parameter selection. Neuroimage 50(2):516–523
Stoilescu L, Maas MC, Huisman HJ (2017) Feasibility of multireference tissue normalization of T2-weighted prostate MRI. In: Proceedings of the 34th annual scientific meeting, European Society for Magnetic Resonance in Medicine & Biology, Barcelona, p 353
Niaf E, Rouviere O, Lartizien C (2011) Computer-aided diagnosis for prostate cancer detection in the peripheral zone via multisequence MRI. In: Proceedings of SPIE medical imaging, Lake Buena Vista, p 79633P
Niaf E, Rouviere O, Mege-Lechevallier F, Bratan F, Lartizien C (2012) Computer-aided diagnosis of prostate cancer in the peripheral zone using multiparametric MRI. Phys Med Biol 57(12):3833–3851
Engelhard K, Hollenbach HP, Deimling M, Kreckel M, Riedl C (2000) Combination of signal intensity measurements of lesions in the peripheral zone of prostate with MRI and serum PSA level for differentiating benign disease from prostate cancer. Eur Radiol 10(12):1947–1953
Dikaios N, Alkalbani J, Abd-Alazeez M, Sidhu HS, Kirkham A, Ahmed HU, Emberton M, Freeman A, Halligan S, Taylor S, Atkinson D, Punwani S (2015) Zone-specific logistic regression models improve classification of prostate cancer on multi-parametric MRI. Eur Radiol 25(9):2727–2737
Dikaios N, Alkalbani J, Sidhu HS, Fujiwara T, Abd-Alazeez M, Kirkham A, Allen C, Ahmed H, Emberton M, Freeman A, Halligan S, Taylor S, Atkinson D, Punwani S (2015) Logistic regression model for diagnosis of transition zone prostate cancer on multi-parametric MRI. Eur Radiol 25(2):523–532
Dollar P, Appel R, Belongie S, Perona P (2014) Fast feature pyramids for object detection. IEEE Trans Pattern Anal Mach Intell 36(8):1532–1545
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak J, van Ginneken B, Sanchez CI (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88
Litjens G, Toth R, van de Ven W, Hoeks C, Kerkstra S, van Ginneken B, Vincent G, Guillard G, Birbeck N, Zhang J, Strand R, Malmberg F, Ou Y, Davatzikos C, Kirschner M, Jung F, Yuan J, Qiu W, Gao Q, Edwards PE, Maan B, van der Heijden F, Ghose S, Mitra J, Dowling J, Barratt D, Huisman H, Madabhushi A (2014) Evaluation of prostate segmentation algorithms for MRI: the PROMISE12 challenge. Med Image Anal 18(2):359–373
Armato SG 3rd, Huisman H, Drukker K, Hadjiiski L, Kirby JS, Petrick N, Redmond G, Giger ML, Cha K, Mamonov A, Kalpathy-Cramer J, Farahani K (2018) PROSTATEx Challenges for computerized classification of prostate lesions from multiparametric magnetic resonance images. J Med Imaging (Bellingham) 5(4):044501
Tustison NJ, Avants BB, Cook PA, Zheng YJ, Egan A, Yushkevich PA, Gee JC (2010) N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 29(6):1310–1320
Otsu N (1979) Threshold selection method from gray-level histograms. IEEE Trans Sys Man Cybern 9(1):62–66
Bojorquez JZ, Bricq S, Brunotte F, Walker PM, Lalande A (2016) A novel alternative to classify tissues from T 1 and T 2 relaxation times for prostate MRI. Magn Reson Mater Phys 29(5):777–788
Delong ER, Delong DM, Clarkepearson DI (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44(3):837–845
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc B 57(1):289–300
Stoyanova R, Takhar M, Tschudi Y, Ford JC, Solorzano G, Erho N, Balagurunathan Y, Punnen S, Davicioni E, Gillies RJ, Pollack A (2016) Prostate cancer radiomics and the promise of radiogenomics. Transl Cancer Res 5(4):432–447
Vos PC, Hambrock T, Barenstz JO, Huisman HJ (2010) Computer-assisted analysis of peripheral zone prostate lesions using T2-weighted and dynamic contrast enhanced T1-weighted MRI. Phys Med Biol 55(6):1719–1734
Acknowledgements
Open Access funding provided by NTNU Norwegian University of Science and Technology (incl St. Olavs Hospital - Trondheim University Hospital). We would like to thank Radboud University Nijmegen in addition to the organizers of the PROSTATEx and PROMISE12 challenges for making their datasets available. We would specifically like to thank Prof. Radka Stoyanova and her team at Miller School of Medicine (Miami, FL, USA) for providing us with the prostate delineations for the PROSTATEx dataset. We would like to thank Dr. Elise Sandsmark from St. Olavs Hospital, Trondheim University Hospital (Trondheim, Norway), for providing us with prostate delineations for the in-house collected dataset.
Funding
The Research Council of Norway (Grant Number 295013), Norwegian University of Science and Technology Biotechnology (Grant Number 81770928), and the liaison Committee between the Central Norway Regional Health Authority and the Norwegian University of Science and Technology (Grant Numbers 90368401 and 90265300).
Author information
Authors and Affiliations
Contributions
Study conception and design were performed by ME and TFB. Material preparation and data collection were performed by MRSS, KMS and ME. Data analysis and interpretation were performed by MRSS, GAK and ME. The first draft of the manuscript was written by MRSS and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in-house in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee [Regional Committee for Medical and Health Research Ethics (REC) in mid Norway, identifier 2017/576] and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sunoqrot, M.R.S., Nketiah, G.A., Selnæs, K.M. et al. Automated reference tissue normalization of T2-weighted MR images of the prostate using object recognition. Magn Reson Mater Phy 34, 309–321 (2021). https://doi.org/10.1007/s10334-020-00871-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10334-020-00871-3