Abstract
Forests are a global environmental priority that need to be monitored frequently and at large scales. Satellite images are a proven useful, free data source for regular global forest monitoring but these images often have missing data in tropical regions due to climate driven persistent cloud cover. Remote sensing and statistical approaches to filling these missing data gaps exist and these can be highly accurate, but any interpolation method results are uncertain and these methods do not provide measures of this uncertainty. We present a new two-step spatial stochastic random forest (SS-RF) method that uses random forest algorithms to construct Beta distributions for interpolating missing data. This method has comparable performance with the traditional remote sensing compositing method, and additionally provides a probability for each interpolated data point. Our results show that the SS-RF method can accurately interpolate missing data and quantify uncertainty and its applicability to the challenge of monitoring forest using free and incomplete satellite imagery data. We propose that there is scope for our SS-RF method to be applied to other big data problems where a measurement of uncertainty is needed in addition to estimates.
Similar content being viewed by others
Introduction
Forest management is a global priority identified by the United Nations and World Bank Sustainable Development Goals (SDGs) [1] and is important in most countries in the world. Satellite images are a freely available and frequently updated data source for forest monitoring. Forest presence can be identified in satellite images for large scale areas by calculating vegetation indices from the spectral bands, which are the colours and near-infrared information captured in the images [2]. Large scale forest cover maps are a transparent tool for identifying forest growth, forest clearing and degradation, and quantifying environmental issues associated with these changes in forest cover such as biodiversity, carbon stocks and social welfare [3]. Examples of these forest cover maps derived from satellite images at global and regional scales include [3,4,5] and [6]. The benefits of using satellite images to construct large scale forest cover maps have been demonstrated for decades in the remote sensing field and they are often the only practical way to monitor forests at regional and global scales [7]. However, a key challenge with using satellite images for forest mapping is missing data due to cloud cover. This is particularly a problem in tropical regions because of the climate driven frequent cloud cover [8]. Unfortunately, these tropical regions of the world with the most persistent cloud cover are where large scale and frequent monitoring is most needed [9] because they are home to large, ecologically important forests, and these forests are continually impacted by seasonal effects and human intervention such as land clearing [10]. The missing data problem occurs for all spectral satellites, including Landsat 7 and Landsat 8, and for newer satellites such as Sentinel 2 [11]. Radar satellites exist which can capture data even beneath clouds, but these images are noisy and do not capture spectral information needed to derive vegetation indices [9], such as Normalised Differenced Vegetation Index (NDVI) and Enhanced Vegetation Index (EVI), which are commonly used for crop and forest detection and monitoring. Regardless of technology improvements to new satellites, spectral satellite images which have this cloud cover missing data problem are currently still the best available free data for monitoring forests at large scales. This means we continue to need methods for dealing with the cloud cover missing data problem. In the remote sensing field, the traditional approach to this issue is interpolation via compositing, which uses the most recently observed values of the pixels from previous time points to fill the missing gaps in the current image [12]. This approach can be successful when there is minimal change in the land cover between images and when the missing pixels have been observed very recently. A limitation of the compositing approach is that it does not account for uncertainty. Any interpolation of data will have some uncertainty, and we need ways to quantify this to obtain more statistically complete estimates and inferences. In this paper we present a new stochastic spatial random forest (SS-RF) method that addresses both of the gaps identified above and builds on the idea of compositing by using recently observed values of the missing data to produce an interpolated land cover classification. We cast this approach in a Bayesian framework to provide posterior probabilistic updates of the required classifications. This has a novel additional benefit of providing a measure of uncertainty attached to the interpolated values. In addition to compositing, there are statistical approaches to interpolating missing data in satellite images including canonical spatial kriging-based approaches [13, 14] spectral-based and temporal-based models, and hybrid combinations of these [15]. In previous work [16] we developed a machine learning approach to spatial interpolation which is fast and accurate. Machine learning methods are popular and useful for remote sensing applications including for identifying clouds and cloud shadow as a pre-processing step to interpolation [11, 17], and monitoring changes in forest cover [18]. Machine learning algorithms are also popular for important environmental monitoring beyond remote sensing applications including identifying deforestation [19] and landslide susceptibility [20].
There are many types of machine learning methods in the literature. We chose a decision tree method, random forest, for our study model because these models are flexible, non-parametric, scalable to big data problems [21], and have been popular in the remote sensing field for over two decades due to their high accuracy in performing satellite image classification [22,23,24].
In our machine learning approach to geostatistical interpolation we used decision tree methods, spatial information and observed sections of an image to train methods and interpolate missing data in the same image at the same point in time. That is, we trained and tested a random forest models on the spectral information that was captured by the satellite, then used these models to interpolate the values of pixels which were missing due to simulated clouds. Importantly, we utilised only latitude and longitude as covariates in the random forest model. This is the minimal information available for all images even when spectral data are missing. We demonstrated that this approach was surprisingly accurate [16]. However, like traditional compositing, all these statistical approaches to spatially interpolating missing data also do not measure the uncertainty of the interpolated values they produce.
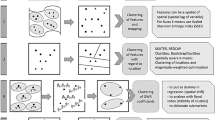
Satellite images can be considered as space-time data [25] because they provide repeated measures, in terms of spectral information, as the same spatial locations at multiple points in time. Our SS-RF method uses images of the same area from recent points in time that have no or minimal missing data to provide a probabilistic statistical solution for filling data gaps in images with most or all of the spectral data missing. The outputs of our method are a probability and an interpolated value. As an overview, our method fits a random forest with spatial information [16] to observed satellite images of the same area captured on previous, recent dates and constructs a Beta distribution to produce both a land cover classification and a probability of the classification of interest. The values from the Beta distribution can be used to interpolate missing data in images and provide a measurement of uncertainty of the interpolated values. We demonstrate our new SS-RF method in a case study interpolating forest presence in regions of satellite images where spectral data are missing due to simulated cloud cover. We produce spatial plots of the probabilities produced by the SS-RF, which identify regions of high and low uncertainty about the presence of forest cover, in addition to classifications and an associated probability for each observation. The paper is structured as follows. In “Materials and methods” section we introduce the case study data, steps taken to calculate vegetation indices from the satellite images, and simulate clouds. We then describe the SS-RF method in detail. In “Results” section we present results of our SS-RF method and compare these with results from compositing, the traditional remote sensing interpolation approach. This section is followed by a discussion of the results, including comparisons between the SS-RF method and compositing, benefits and limitations of the SS-RF approach and future work in “Discussion” section. Finally, we summarise our method and results in “Conclusion” section.
Materials and methods
Study area
The case study area is in Queensland, Australia, approximately 100 km North West of the Injune township. The 37 × 60 km landscape in this region is comprised of forest, pasture and agricultural land [26] (Fig. 1). For this site we have a time series of Landsat 8 images, captured at 16 day intervals between 24 July 2017 to 27 July 2018. For the images that are not missing due to cloud cover we can calculate our variable of interest which is Normalised Differenced Vegetation Index (NDVI). This is described in the target response subsection. As a vegetation index, NDVI can theoretically range from − 1 to 1, with values closer to 1 indicating forest and vegetation and values below 0 generally indicating areas without vegetation including bare earth, water or snow. In the case study data NDVI is generally within the range from 0 to 1 and missing data are NA. There are a number of images that weren’t provided in the time series, which leaves 22 images which can be processed, and of these some regions in the images are missing due to cloud, as shown by the white areas in Fig. 2.
In Fig. 2 the darker green regions indicate forest land cover and lighter green indicate not forest land cover. White regions indicate the data are missing due to cloud cover or other atmospheric effects. In some cases, such as 31/12/2017 where the entire plot is white, the entire area in the image is missing due to cloud cover.
Calculating response variables: vegetation index and binary land cover
The variable we use to identify forest presence and absence is Normalised Differenced Vegetation Index (NDVI). This variable is a measure of greenness which can be used to identify the presence and different types of vegetation in the landscape [27]. We calculate NDVI based on Landsat 8 satellite images using the remote sensing formula
where r is the ratio of reflectance in the two spectral bands recording the colour red and near infrared (NIR) information in the satellite image [28]. We identify forest using NDVI as the measure of vegetation because it is widely used and can be calculated easily from a freely available satellite image. Based on a cut-off value of \(0 \le 0.4\) not forest and \(0.4 \ge 1\) forest [27] we produce a binary land cover classification identifying pixels as forest or not forest.
Spatial scales
We perform analysis at two spatial scales. The first is at the original satellite image scale which is pixel level. A Landsat 8 pixel is approximately 30 m \(\times\) 30 m [26]. We also perform analysis at a more aggregated ‘neighbourhood’ scale. The intention is to explore whether the performance of the SS-RF method is impacted by data at different spatial scales because spectral satellite images are available at different resolutions e.g. 30 m for Landsat and 250 m for MODIS satellites [29]. To create neighbourhoods we aggregate pixels within the same area and assign the neighbourhoods a value of the mean NDVI value of all the pixels in that neighbourhood. We specify each neighbourhood to have 33 \(\times\) 33 pixels as per Colin et al (2018) [30] using the Raster package [31] in R [32]. An example of NDVI at neighbourhood scale is shown in Fig. 3. Aggregating the NDVI data to neighbourhood scale produces a smoother image than data at pixel scale (Fig. 3). We use the SS-RF method on both spatial scales to predict the probability of pixel land cover being forest, and the probability of the neighbourhood land cover being forest.
Image selection and simulating missing data
In the SS-RF method we interpolate simulated missing data in an image by using data from images of the same area at two recent points in time when the data were observed. We selected four images which were observed in the time series, which we refer to as \(t-1\), t, \(t+1\) and \(t+2\). In order to assess the performance of the SS-RF method we selected images with no missing data and then simulated missing data using our approach we describe in this section. We selected only observed images (with no missing data) because in order to assess the performance of the SS-RF method at interpolating missing data, we need to know the real values of the pixels and neighbourhoods. Based on these requirements, the time points chosen are:
-
\(t+2\): 10 September 2017
-
\(t+1\): 25 August 2017
-
t: 9 August 2017
-
\(t-1\): 24 July 2017
For the SS-RF method we use images at \(t-1\) and t to interpolate data in image \(t+1\), and use images at t and \(t+1\) to interpolate data in image \(t+2\). For the compositing method, we use only the most recent observed image to interpolate the missing data. We use the values of image t to interpolate \(t+1\) and values of \(t+1\) to interpolate \(t+2\). We simulated clouds to mimic how missing data appears in satellite images and retained the true values to validate the results of the SS-RF and compositing methods. In order to produce our own missing data, we simulated missing data in cloud patterns on the images \(t+1\) and \(t+2\). We simulated the missing data patterns independently of the NDVI based forest presence data using the process described in [16] based on [33]. We applied the SS-RF method to the same pixels and neighbourhoods in each image, identified by their geographical location (longitude and latitude), to ensure we were examining the same observations over time and could make fair assessments of model performance when interpolating the values at future time points \(t+1\) and \(t+2\).
Figures 4 and 5 show the original NDVI images as a binary land cover plot of the areas with forest indicated by green and not forest indicated by grey. The land cover data in Fig. 4 are at pixel scale which is the original, finer resolution. The differences in forest cover between areas are more distinct at this spatial scale (Fig. 5).
At both pixel and neighbourhood scales there is a difference in the composition of land cover between areas 1 and 2; area 1 has more forest cover than area 2 which is predominantly not forest at the image dates chosen. We selected these images deliberately to examine the performance of the model on different land cover compositions; one image which is predominantly one land cover class (area 2) and one which is a more heterogeneous combination of the two land cover classes (area 1).
General SS-RF method
Our SS-RF method uses a Bayesian recursive estimation scheme based on Beta distributions to represent the probability of a pixel or neighbourhood in an image belonging to one of two land cover classifications. We use the Beta distribution because in our case study we are interested in a binary variable; forest or not forest land cover. Let \(I^{(t)}\) denote the image at time t and \(P_i^{(t)}\) denote the probability of forest in the jth region of this image. Assume that \(P_i^{(t)}\) follows a Beta distribution,
The SS-RF scheme updates the hyper-parameters \(\alpha _i^{(t)}\) and \(\beta _i^{(t)}\) which can be expressed as
where \(v_i^t\) is the corresponding variance of the Beta distribution. The SS-RF scheme proceeds by taking the image \(I^{(t)}\) at time t as a prior for the image \(I^{(t+1)}\) at time \(t+1\). Hence by Bayes’ theorem [34], the posterior probability of forest in the ith region of image \(I^{(t+1)}\) is given by
with posterior mean given by
In the SS-RF scheme, the values of p at times t and \(t-1\) are obtained using a random forest (RF) algorithm. The RF algorithm is a machine learning method that classifies observations based on the average of many shallow decision trees, each of which is constructed using a random sample of the regions in the image [21]. This so-called bagging approach, combined with the use of training and test sets to respectively train each decision tree and evaluate its performance, produces highly accurate classifications and predictions [35]. The values of v are specified based on the variance observed in the testing and training samples across the images.
In our case study we draw values of each observation from the beta distribution and treat these as the values of the observations that are missing due to cloud. The sequential steps in our algorithm are outlined in Appendix A.
SS-RF method applied to case study
In this case study we calculated the variance in NDVI observed in the random sample used for testing and training across the images. Based on the results we set the value for variance v in Eq. (1) to 0.01 for the image at time t and allowed a larger value of 0.02 for the older image \(t-1\) (see Appendix C). We predicted the probability of the land cover class being forest at both pixel and neighbourhood spatial scales as a function of latitude and longitude of the neighbourhoods per [16]. We take a sub sample of 100,000 observations from both the images at previous time points, t and \(t-1\). We split these into training samples of 80,000 and testing samples of 20,000, which represents an 80/20 per cent split. We train the random forests on the training samples, and produce probabilities of land cover being forest for the testing sets. We then apply the trained models to an additional random subset of the data of 200,000 observations to produce probabilities of land cover being forest. We then take these probabilities for the previous images, t and \(t-1\), and use these in Eq. (3) to generate the Beta distribution, \(E(p_i^{(t)})\). The expected values of this Beta distribution then gives our interpolated probabilities of forest cover at \(t+1\) and uncertainty of these posterior predictions which are missing due to simulated cloud cover.
That is, we infer the probability of observations at \(t+1\) as being the probability of the observations being forest at time t, given the probability of being forest at time \(t-1\). These posterior probabilities, inform the decision rule which determines the class k of neighbourhood i at the future time point \(t+1\) as forest or not forest.
For each sample of data we obtained a probability of each observation being forest and a binary classification as forest or not forest where the former is based on the expectation of the posterior probability distribution and the latter is based on whether this expected value exceeds the threshold of 0.5. In our case study where we have the true observed land cover classes that are underneath the simulated clouds, we compare the plotted posterior probability of forest with the true land cover classes to evaluate how well the model can interpolate the land cover. We assessed the accuracy of the SS-RF method at interpolating missing data by producing confusion matrices using the true class values of the testing data set and the predicted probabilities performed on the testing data set converted to forest or not forest classifications.
To examine the performance of the SS-RF method compared to the standard remote sensing solution to interpolating missing data, we performed compositing and compared the results with the SS-RF outputs. Compositing interpolates missing image data by taking the most recently observed value of each pixel and using this as the true value for missing data. It is an absolute value and no uncertainty is included in this approach. To perform compositing we used the most recently observed pixel values to interpolate the simulated missing data at \(t+1\) and \(t+2\). Specifically, for \(t+2\) we interpolated the missing data with the values for the same pixels and neighbourhoods at \(t+1\), and interpolated the data at \(t+1\) using data from t. We produced confusion matrices of the true class values of the testing data sets and the interpolated values produced by compositing. We implemented the SS-RF and compositing methods for both the study areas 1 and 2 at pixel and neighbourhood scale, then compared the accuracy of the interpolation results produced by the SS-RF method and compositing.
Results
We first present an example of the full outputs of the SS-RF method. The outputs take the form of spatial plots of interpolated values identifying how probabilities of forest are distributed, and plots of interpolated land cover classifications. Secondly, we present the results of the SS-RF method and make comparisons between the two study areas, and results for pixel and neighbourhood scale. Thirdly, we present a comparison of the results of the SS-RF and compositing methods for area 1 and area 2 at pixel and neighbourhood scale. Finally, we present the compositing results and make comparisons between study areas and pixel and neighbourhood scale.
Visualising probabilities of forest land cover
The SS-RF method outputs can be used to effectively visualise the spatial distribution of both discrete land cover classifications and posterior probabilities of forest in the area of interest (Fig. 6). These visualisations provide maps of interpolated forest cover data, and the uncertainty of those data values. In Fig. 6 the posterior probability of forest as predicted by the SS-RF method is plotted by latitude and longitude. This can be viewed as an uncertainty map of the interpolated forest cover data (Fig. 6). In Fig. 7 the true class values observed before we simulated clouds for the sample and area are plotted by latitude and longitude. By comparing true land cover classes (forest and not forest) with the probabilities, we can see that the SS-RF method accurately identifies land cover for this sample (Fig. 7). For example, in the top grid of Figure 7 between 147.3 and 147.4 on the x axis, there is a region of predominantly grey observations which indicates that the land cover is not forest. These same observations in Fig. 6 are predominantly dark blue, which indicates low probability of forest. There is concordance between the posterior probabilities and true class values of these points (Fig. 8). Fig. 8 shows the interpolated land cover classifications; forest and not forest, that were produced for the same sample of observations shown in Figs. 6, 7 using the SS-RF method. Figure 8 shows the probabilities produced by the SS-RF method for sample 5 of area 1 on 10 September 2017 plotted by latitude and longitude as a binary land cover classification. If the probability of the observation being forest is equal to or greater than 0.50, the observation is plotted as green and if the probability of the observation being forest is less than 0.5, it is plotted as grey. We observe the same pattern as in Fig. 6, with predominantly grey ‘not forest’ land cover observed in the top grid between 147.3 and 147.4 longitude.
Posterior probability of forest at pixel scale obtained from SS-RF method plotted by spatial coordinates. The probability that each pixel has a land cover classification of forest is indicated by the shading; darker blue indicates low probability the pixel is forest and light blue indicates high probability of forest in that pixel. The pixels are in patches because they are part of a testing data sample that was taken at random within the clouded area of the image
SS-RF method results
The accuracy of the SS-RF method for interpolating land cover data was higher for area 2 than for area 1 across both resolutions and image dates (Fig. 9). Overall accuracy for area 2 was higher for neighbourhood scale data, ranging from 0.955 to 0.967. Overall accuracy for pixel scale data was 0.929 to 0.939. For area 2 overall accuracy was slightly higher for 10 September 2017 image than the 25 August 2017 image, for both pixel and neighbourhood scale data (Fig. 9). The accuracy of the SS-RF for interpolating land cover data for area 1 was more variable than for area 2. This is expected as area 1 had a more heterogeneous structure of the two land cover classes forest and not forest. The SS-RF was most accurate predicting land cover at pixel scale, with accuracy ranging from 0.749 to 0.831. Overall accuracy for neighbourhood scale data ranged from 0.408 to 0.656, with 0.408 appearing as an outlier in the results for the neighbourhood scale data and in the overall results for area 1 (Fig. 9). The low accuracy for this sample of data seems to be due to the balance of the classes in the training data set compared with the values in the testing data set. There were 11,343 (56.7%) forest and 8657 (43.3%) not forest land cover observations in the training set and in the testing set that was predicted there were 50,271 (63.8%) forest and 29,729 (37.2%) not forest land cover observations. For the 25 August 2017 image model performance was most accurate for pixel scale data with overall accuracy ranging from 0.825 to 0.831. For the 10 September 2017 image, model performance was most accurate for pixel scale data with overall accuracy ranging from 0.749 to 0.760.
Comparison of SS-RF method and compositing interpolation
Importantly, we found the SS-RF method had accuracy comparable with the compositing method in most situations. In situations where the SS-RF method was not as accurate it had the additional benefit of providing the probability of forest presence in addition to a classification. The relative performance of the SS-RF method compared with compositing varied across resolutions and the two areas.
For area 2, overall the performance of the SS-RF method and compositing was comparable at both neighbourhood and pixel scales, as shown in Fig. 10. For the 25 August 2017 image (t) overall accuracy of the SS-RF method across both resolutions ranged from 0.929 to 0.960, which is comparable with 0.968 to 0.971 for compositing. For the 10 September 2017 image overall accuracy of the SS-RF method ranged from 0.932 to 0.967 and from 0.966 to 0.985 for compositing.
Figure 11 shows model performance for area 1, which is the more heterogeneous of the two areas in terms of combination of forest and not forest land cover and less stable in terms of change over time. For area 1 the compositing approach had higher overall accuracy than the SS-RF method, at both neighbourhood and pixel scales. The difference in performance between the two methods was more obvious at neighbourhood scale, with the SS-RF method producing less accurate results below 0.65 for both image dates, and had particularly low accuracy for the 25 August 2017 image. At pixel scale this difference in performance between methods was slightly reduced, with SS-RF overall accuracy above 0.75 for both image dates, and above 0.82 for 25 August 2017.
Compositing results
Using a compositing approach, for 10 September 2017 (\(t+2\)) missing data we interpolated using the most recently observed data, which were from 25 August 2017 (\(t+1\)). For the 25 August 2017 image (\(t+1\)) we interpolated using the most recently observed data from the previous satellite image on 9 August 2017 (t). The accuracy of compositing for interpolating land cover data was slightly higher for area 2 across both resolutions and image dates (Fig. 12). Overall accuracy for area 2 was higher for neighbourhood scale data, ranging from 0.971 to 0.985. Overall accuracy for pixel scale data ranged from 0.966 to 0.970. Overall for both images, compositing produced highly accurate results for missing data interpolation.
Discussion
Model accuracy
Our results show the SS-RF method performs extremely well, with between 93% and 97% overall accuracy, when the land cover data are relatively stable over time and landscape is more homogeneous. This was the case in area 2 for all four images from times \(t-1\) to \(t+2\). Accuracy at pixel scale was high for the heterogeneous land cover data in the 25 August 2017 image (0.825% to 0.831%), however this accuracy was noticeably lower for neighbourhood scale data for the same image (0.408% to 0.534%).
The lower accuracy of the SS-RF method in some cases compared with compositing is driven by changes in the land cover data between the dates of the input images and the image being interpolated. To demonstrate this, we present an example of a sample of data from area 1. The input random forest algorithms that are used to produce the probabilities of forest cover p which are part of the calculation of the parameters \(\alpha\) and\(\beta\) have extremely high accuracy for 9 August and 25 August 2017 between 0.98 and 0.99 (See Appendix D for full results for this sample). The lower accuracy of the SS-RF model is therefore unlikely to be driven by this component of the method, but rather by the following Beta distribution step that relies on observed land cover from two images (Fig. 13). As shown in Fig. 13, the amounts of forest and not forest observations are relatively consistent between the first two images on 9 August 2017 and 25 August 2017, but changes noticeably to more forest cover between 25 August 2017 and 10 September 2017. It appears the lower accuracy of the SS-RF method for 10 September 2017 is based on the fact the land cover observed in the previous two time points is not an accurate representation of the land cover on 10 September 2017.
As another example of the influence of the observed land cover in previous images and stability of the land cover over time we show a case where the SS-RF method had high accuracy (Fig. 14).
Plot of the true land cover classifications for the same sample of neighbourhoods in Area 2 across three time points, plotted by their latitude and longitude. The points are jittered by 0.1 to make interpretation easier as they are spatially close together. Forest is shown in green and not forest in grey
In this case the SS-RF had comparable performance with compositing, driven by the fact that the land cover across all three time points was primarily not forest consistently across the three time points (Fig. 14). Based on these results, we conclude the accuracy of the SS-RF method is influenced by the level of stability in land cover over time, as it performed well when the land cover had minimal change between the observed images and the image being interpolated, and performed poorly when there was more variation in the land cover over time (Fig. 13). We note this limitation is not confined to the SS-RF method, as the accuracy of compositing is also impacted by instability in land cover over time.
SS-RF outputs and method benefits
Our key model output is the posterior Beta distribution which provides the probability of each individual observation belonging to a land cover class, in this case forest or not forest. From this output we progressed to a binary classification; however, the probabilities in their original form could be used to inform forest monitoring and provide a probability of forest presence. In addition to using the probabilities to interpolate the missing data in images, they could also be used to identify areas where the chance of forest presence has decreased or increased and inform decisions about where it could be beneficial to invest in field data collection. The key benefit of our SS-RF method is the ability to produce a measurement of uncertainty for land cover classifications of missing data (pixels or larger geographic regions). These probabilities of forest cover can be used to produce spatial maps of uncertainty to augment the interpolated land cover classifications. Additional benefits of our SS-RF method include its speed, taking only minutes to train and produce results for hundreds of thousands of observations, ability to handle very large amounts of data efficiently, more relaxed statistical assumptions [35] and ease of implementation in open source software R [32]. The method can be easily updated with new data in a recursive manner, which further increases the computational efficiency and scalability of the approach and improves its utility. This is highly relevant to our application where it is of interest to interpolate forest cover in satellite images, and do this for new images over time to monitor forest. Beyond our satellite image case study, the SS-RF method can be applied to other fields, such as biomedicine, which requires binary classification of images and uncertainty measurements for these classifications [36]. Indeed, our method could be applied to any big data classification problem where the user wants to predict the probability of an event or feature of interest. Examples of where this is useful include predicting the probability of disease in health, species presence in ecology, and crops in agricultural monitoring.
Comparing SS-RF interpolation with compositing
In the case study, the SS-RF method performed inconsistently compared with compositing across the two different areas. For area 2, the SS-RF method results were comparable with those for compositing at both neighbourhood and pixel scale. For area 1 in our case study the compositing results were more accurate than the SS-RF land cover predictions at both neighbourhood and pixel scale. We used overall accuracy produced from confusion matrices to compare the SS-RF method and compositing because it is possible to produce a classification from both approaches, however it is not possible to produce a probability from compositing. This means that the benefit of having a probability of forest cover, which is a key contribution of our method, was not directly incorporated into the comparison metric. Since the compositing method assumes the most recently observed values of the pixels in images, for example at \(t-1\) and t, are the same, it implicitly assumes a 100% probability of the pixels having the same land cover classification at both points in time. This is unlikely to be true for all observations, and compositing does not account for this uncertainty whereas the SS-RF method does. The probabilities for the interpolated land cover produced by the SS-RF give this method an advantage over compositing overall.
We were able to assess the accuracy of the compositing approach compared with our SS-RF method in our case study because we simulated our clouds and missing data. However, in real cases of missing data in images the true values of the data are unknown. Although the SS-RF method did not consistently match the accuracy of the traditional compositing approach, using the SS-RF method to interpolate missing data is beneficial because it provides an uncertainty measurement in addition to a land cover classification. It could be beneficial to use a combination of compositing and SS-RF probabilities to interpolate missing data i.e. using the most recently observed values to create a composite as a baseline and highlighting observations where the SS-RF and composite values differ. For example, if according to the compositing approach an observation is not forest and according to the SS-RF method the observation is forest. A limitation of both compositing and the SS-RF method is reliance on stability in land cover over time. When a significant change in land cover occurs between images, both compositing and the SS-RF method will have lower accuracy interpolating the missing data. Where the SS-RF method has an advantage in this scenario is the ability to quantify the uncertainty of its land cover predictions, whereas compositing only provides a land cover prediction based on the most recently observed data.
Our case study was an example where there were four consecutive images and we were interpolating data at the next future time point e.g. \(t+1\) using t and \(t-1\). However, in cases where you want to interpolate an intermediate step e.g. you have an observed image at time t and at time t +2 and want to interpolate \(t+1\) under a compositing approach it is unclear which data is preferable to use. The SS-RF method does not require a choice between the observed data, but uses data from both observed images to inform predictions.
Thresholds for land cover classifications and limitations
To produce land cover classifications from the probability values produced by the SS-RF method we used a probability threshold of 0.5 to classify observations as forest or not forest. Under this threshold, observations with probabilities 0.5 or greater are classified as forest and not forest otherwise. When probabilities are close to the extreme values 0 and 1 this is a clear rule, however for probabilities around 0.5 there is more uncertainty around which is the appropriate land cover class. For other case studies different thresholds may be selected based on a different loss or objective function, or using an ROC curve to inform threshold choice. Regardless of the application, the selection of an appropriate threshold is a challenging and subjective task.
A limitation is that the SS-RF method cannot detect sudden changes, sometimes referred to as change points, in the landscape. See for example Fig. 13 where there was a change in the true classes between time t and \(t+1\). There is likely to be a lag in detecting such change in the images over time. For example, if there had been a dramatic event such as tree clearing during the 16 days between satellite images and this land cover change was obscured by cloud, the model would predict forest presence similar to the previous observations. At least one time step of observed data is needed that reflects the forest cover change to start to incorporate any such sudden changes into the predictions.
Although the SS-RF method uses images from different points in time, it is not a time series method. The method does account for some effects of time by imposing a larger variance for the older image to give more weight to the influence of the most recent image when performing the interpolation. The treatment of data which are temporally distant as though they are captured at the same point in time is not unusual in satellite image processing. For example, the common practice of compositing we demonstrated in this paper substitutes missing pixels with observed pixels of the same region from a different time point and treats the pixels in the composited image as being from the same time point. Our method extends on compositing by using the most recently observed images of the same area to produce a probability of an observation belonging to a land cover class, in addition to a land cover classification.
Extensions
We selected a decision tree method, random forest, for our study model because this method offers key benefits for big data problems as described in the introduction. Other supervised classification methods that are currently popular for satellite image analysis include support vector machine [37,38,39] and deep learning methods: convolutional neural networks (CNNs) [40, 41] and generative adversarial networks (GANs) [42]. These methods could potentially be implemented within our stochastic framework in future studies.
In this case study we focused on identifying two classes; our land cover of interest; forest, and all other land cover was classified as not forest. This binary example necessitated the use of the Beta distribution, which by definition is binary. To extend the SS-RF method to more than two classes, a Dirichlet distribution could be implemented as an alternative. In cases where there are multiple images in a row with missing data the outputs of the SS-RF method could be used in combination with observed data to interpolate missing data at a future time. For example, if the images at \(t-1\) and t are observed and used to interpolate \(t+1\) but \(t+2\) also has missing data, the interpolated values of \(t+1\) could be used with the observed values at t to interpolate \(t+2\), and continuing in this Bayesian updating scheme using estimated land cover data until data are observed. The benefit of this approach is that having long periods of cloud cover, as is common in tropical areas, will not prevent the use of satellite images for forest monitoring and the uncertainty of the interpolated data is measured. ePrevious images of the same area were used in this case study because these are the data we have; the fact they are a sequence of images in time is not essential to the method. This approach could also be used for cases with multiple images of the same area at the same point in time i.e. at different resolutions or from different data sources (aerial photograph, satellite image).
In our case study we simulated clouds so we knew the true values of observations at \(t+1\) and \(t+2\), and hence we were able to assess model accuracy. In cases where the cloud data are truly missing, predicting values in images ahead of the image being taken or available for analysis this is not possible. This is where the probabilistic nature of the method is useful. In the case of truly missing data the real land cover values under the clouds are unknown, therefore having an informed probability of the observations being forest, in addition to a land cover classification is helpful. Having land cover classifications, and a measurement of the uncertainty around these, could inform forest management planning by identifying forest cover or predicting trajectories of natural regeneration or deliberate reforestation.
Conclusion
Forest management is a priority identified by the United Nations and World Bank Sustainable Development Goals (SDGs) [1] and is important globally. Satellite images are a freely available and frequently updated data source for forest monitoring. However, missing data due to frequent cloud cover is a main barrier to using these data, particularly in tropical areas where forest monitoring is essential. We have extended our previous spatial decision tree approach to image interpolation [16] and presented our new SS-RF method to statistically predict missing data in satellite images, and importantly, provide a measurement of uncertainty for these predictions.
The SS-RF method is fast, scalable to large amounts of data and accurate, performing probabilistic binary land cover prediction with overall accuracy up to 0.97. We compared the performance of our new method with the remote sensing standard approach; compositing. We found when land cover is relatively stable over time the SS-RF method produced comparable results to compositing for predicting binary land cover; forest and not forest, with the additional benefit of producing a probability of forest cover as well as a land cover classification. In cases where the SS-RF method did not perform as well as compositing, the probabilities it produced were useful to provide a measure of uncertainty for the interpolated values. In a real scenario where data are truly missing, it is not possible to know whether compositing or any method is accurate or not, which makes having a probability associated with interpolate values so essential. This is the key benefit of the SS-RF method. Beyond our satellite image case study, the SS-RF method can be applied to other large images, such as those used in medicine or indeed any big data problem where the user wants to predict the probability of an event or feature of interest.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- SS-RF::
-
Spatial Stochastic Random Forest
- NDVI::
-
Normalised Differenced Vegetation Index
References
United Nations. Global indicator framework for the Sustainable Development Goals and targets of the 2030 Agenda for Sustainable Development. Tech. rep., United Nations; 2018. https://unstats.un.org/sdgs.
Gibson P, Power C. Introductory remote sensing principles and concepts. London: Routledge; 2013. https://doi.org/10.4324/9780203714522.
Hansen MC, Potapov PV, Moore R, Hancher M, Turubanova SA, Tyukavina A, Thau D, Stehman SV, Goetz SJ, Loveland TR, Kommareddy A, Egorov A, Chini L, Justice CO, Townshend JRG. High-resolution global maps of 21st-century forest cover change. Science. 2013;342(6160):850–3. https://doi.org/10.1126/science.1244693.
Global Forest Watch. Interactive map: global forest watch; 2020. https://www.globalforestwatch.org/map/.
Liu L, Tang H, Caccetta P, Lehmann EA, Hu Y, Wu X. Mapping afforestation and deforestation from 1974 to 2012 using Landsat time-series stacks in Yulin District, a key region of the Three-North Shelter region, China. Environ Monit Assess. 2013;185(12):9949–65. https://doi.org/10.1007/s10661-013-3304-2.
Echeverria C, Coomes D, Salas J, Rey-Benayas JM, Lara A, Newton A. Rapid deforestation and fragmentation of Chilean temperate forests. Biol Conserv. 2006;130(4):481–94. https://doi.org/10.1016/j.biocon.2006.01.017.
Bullock EL, Woodcock CE, Olofsson P. Monitoring tropical forest degradation using spectral unmixing and Landsat time series analysis. Remote Sens Environ. 2018; 110968. https://doi.org/10.1016/j.rse.2018.11.011. https://linkinghub.elsevier.com/retrieve/pii/S0034425718305200.
Zhu X, Helmer EH. An automatic method for screening clouds and cloud shadows in optical satellite image time series in cloudy regions. Remote Sens Environ. 2018;214:135–53. https://doi.org/10.1016/j.rse.2018.05.024.
Asner GP. Cloud cover in Landsat observations of the Brazilian Amazon. Int J Remote Sens. 2001;22(18):3855–62. https://doi.org/10.1080/01431160010006926. http://www.tandfonline.com/action/journalInformation?journalCode=tres20.
Asner GP, Knapp DE, Broadbent EN, Oliveira PJC, Keller M, Silva JN. Selective Logging in the Brazilian Amazon. Science. 2005;310:480–2. https://doi.org/10.1126/science.1118051, http://www.sciencemag.org/cgi/content/full/310/5747/480/.
Singh P, Komodakis N. Cloud-Gan: Cloud Removal for Sentinel-2 Imagery Using a Cyclic Consistent Generative Adversarial Networks. In: IGARSS 2018 - 2018 IEEE international geoscience and remote sensing symposium, IEEE, Valencia, Spain; 2018. p. 1772–1775. https://doi.org/10.1109/IGARSS.2018.8519033. https://ieeexplore.ieee.org/document/8519033/.
Flood N. Seasonal Composite Landsat TM/ETM+ images using the medoid (a Multi-Dimensional Median). Remote Sens. 2013;5(12):6481–500. https://doi.org/10.3390/rs5126481. https://www.mdpi.com/2072-4292/5/12/6481.
Pringle MJ, Schmidt M, Muir JS. Geostatistical interpolation of SLC-off Landsat ETM+ images. ISPRS J Photogramm Remote Sens. 2009;64:654–64. http://www.elsevier.com/copyright.
Zhang C, Li W, Travis D. Gaps-fill of SLC-off Landsat ETM+ satellite approach. Int J Remote Sens. 2007;28(22):5103–22. https://doi.org/10.1080/01431160701250416.
Shen H, Li X, Cheng Q, Zeng C, Yang G, Li H, Zhang L. Missing information reconstruction of remote sensing data: a technical review. IEEE Geosci Remote Sens Mag. 2015;3(3):61–85. https://doi.org/10.1109/MGRS.2015.2441912. http://ieeexplore.ieee.org/document/7284768/.
Holloway J, Helmstedt KJ, Mengersen K, Schmidt M. A decision tree approach for spatially interpolating missing land cover data and classifying satellite images. Remote Sens. 2019;11(15):1796. https://doi.org/10.3390/rs11151796. https://www.mdpi.com/2072-4292/11/15/1796.
Shendryk Y, Rist Y, Ticehurst C. Deep learning for multi-modal classification of cloud, shadow and land cover scenes in PlanetScope and Sentinel-2 imagery. ISPRS J Photogramm Remote Sens. 2019. https://doi.org/10.1016/j.isprsjprs.2019.08.018.
de Bem P, de Carvalho Junior O, Fontes Guimarães R, Trancoso Gomes R. Change detection of deforestation in the Brazilian Amazon using Landsat data and convolutional neural networks. Remote Sens. 2020;12(6):901. https://doi.org/10.3390/rs12060901. https://www.mdpi.com/2072-4292/12/6/901.
Mayfield HJ, Smith C, Gallagher M, Hockings M. Considerations for selecting a machine learning technique for predicting deforestation. Environ Model Softw. 2020. https://doi.org/10.1016/j.envsoft.2020.104741.
Dang VH, Hoang ND, Nguyen LMD, Bui D, Samui P. A novel GIS-based random forest machine algorithm for the spatial prediction of shallow landslide susceptibility. Forests. 2020. https://doi.org/10.3390/f11010118.
Hastie T, Tibshirani R, Friedman J. The elements of statistical learning. 2nd ed. New York: Springer; 2008. https://doi.org/10.1007/978-0-387-84858-7.
Belgiu M, Drăguţ L. Random forest in remote sensing: a review of applications and future directions. ISPRS J Photogramm Remote Sens. 2016;114:24–31. https://doi.org/10.1016/J.ISPRSJPRS.2016.01.011.
Berhane T, Lane C, Wu Q, Autrey B, Anenkhonov O, Chepinoga V, Liu H. Decision-tree, rule-based, and random forest classification of high-resolution multispectral imagery for wetland mapping and inventory. Remote Sens. 2018;10(4):580. https://doi.org/10.3390/rs10040580. http://www.mdpi.com/2072-4292/10/4/580.
Pal M. Random forest classifier for remote sensing classification. In J Remote Sens. 2005;26(1):217–22. https://doi.org/10.1080/01431160412331269698.
Kneib T, Fahrmeir L. Structured additive regression for categorical space-time data: a mixed model approach. Biometrics. 2006;62(1):109–18. https://doi.org/10.1111/j.1541-0420.2005.00392.x.
Schmidt M, Lucas R, Bunting P, Verbesselt J, Armston J. Multi-resolution time series imagery for forest disturbance and regrowth monitoring in Queensland, Australia. Remote Sens Environ. 2015;158:156–68. https://doi.org/10.1016/J.RSE.2014.11.015. https://www.sciencedirect.com/science/article/pii/S003442571400457X.
Sentinel Hub. NDVI (Normalized Difference Vegetation Index); 2019. https://www.sentinel-hub.com/eoproducts/ndvi-normalized-difference-vegetation-index.
Auscover. AusCover Good Practice Guidelines. Tech. rep., AusCover; 2015. http://data.auscover.org.au/xwiki/bin/view/Good+Practice+Handbook/WebHome.
United Nations.Earth Observations for Official Statistics: satellite imagery and geospatial data task team report. Tech. Rep. December, United Nations Satellite Imagery and Geospatial Data Task Team; 2017. https://unstats.un.org/bigdata/taskteams/satellite/UNGWG_Satellite_Task_Team_Report_WhiteCover.pdf.
Colin B, Schmidt M, Clifford S, Woodley A, Mengersen K, Colin B, Schmidt M, Clifford S, Woodley A, Mengersen K. Influence of spatial aggregation on prediction accuracy of green vegetation using boosted regression trees. Remote Sens. 2018;10(8):1260. https://doi.org/10.3390/rs10081260. http://www.mdpi.com/2072-4292/10/8/1260.
Hijmans RJ. Package ’raster’: geographic data analysis and modeling; 2017. https://cran.r-project.org/web/packages/raster/raster.pdf.
Team RC. R: a language and environment for statistical computing; 2017. https://www.r-project.org.
Helmstedt KJ, Potts MD. Valuable habitat and low deforestation can reduce biodiversity gains from development rights markets. J Appl Ecol. 2018;55(4):1692–700. https://doi.org/10.1111/1365-2664.13108.
Gelman A, Carlin JB, Stern HS, Dunson DB, Vehtari A, Rubin DB, Dominici F, Faraway JJ, Tanner M, Zidek J, Smith PJ. Bayesian data analysis. 3rd ed. Boca Raton: CRC Press; 2014. https://doi.org/10.1201/b16018.
Elith J, Leathwick JR, Hastie T. A working guide to boosted regression trees. J Anim Ecol. 2008;77(4):802–13. https://doi.org/10.1111/j.1365-2656.2008.01390.x.
Shin SJ, Wu Y, Zhang HH, Liu Y. Probability-enhanced sufficient dimension reduction for binary classification. Biometrics. 2014;70(3):546–55. https://doi.org/10.1111/biom.12174.
Kranjčić N, Medak D, Župan R, Rezo M. Support vector machine accuracy assessment for extracting green urban areas in towns. Remote Sens. 2019;11(6):655. https://doi.org/10.3390/rs11060655. https://www.mdpi.com/2072-4292/11/6/655.
Ma L, Liu Y, Zhang X, Ye Y, Yin G, Johnson BA. Deep learning in remote sensing applications: a meta-analysis and review. ISPRS J Photogramm Remote Sens. 2019;. https://doi.org/10.1016/j.isprsjprs.2019.04.015.
Maxwell AE, Warner TA. Fang F (2018) Implementation of machine-learning classification in remote sensing: an applied review. Int J Remote Sens. 2018;10(1080/01431161):1433343.
Mahdianpari M, Salehi B, Rezaee M, Mohammadimanesh F, Zhang Y. Very deep convolutional neural networks for complex land cover mapping using multispectral remote sensing imagery. Remote Sens. 2018;. https://doi.org/10.3390/rs10071119.
Tong XY, Xia GS, Lu Q, Shen H, Li S, You S, Zhang L. Land-cover classification with high-resolution remote sensing images using transferable deep models. Remote Sens Environ. 2020;237(111):322. https://doi.org/10.1016/j.rse.2019.111322.
Wei Y, Luo X, Hu L, Peng Y, Feng J. An improved unsupervised representation learning generative adversarial network for remote sensing image scene classification. Remote Sens Lett. 2020;11(6):598–607. https://doi.org/10.1080/2150704X.2020.1746854.
Acknowledgements
The authors thank Drs. Michael Schmidt, Jessica Cameron and Catherine Leigh for early feedback on the project.
Funding
This research was conducted by the Australian Research Council Centre of Excellence for Mathematical and Statistical Frontiers (Project Number CE140100049) and funded by the Australian Government. K. Helmstedt was funded by the ARC Fellowship DE200101791.
Author information
Authors and Affiliations
Contributions
Jacinta Holloway-Brown performed the literature review, contributed to the methodology development, implemented and evaluated the method and drafted the manuscript. Kate Helmstedt contributed to development of concepts and revision of the manuscript. Kerrie Mengersen contributed to the methodology development and revision of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendices
Appendix A: SS-RF algorithm
The steps in our SS-RF algorithm are outlined below.
-
1.
Form prior from image \(I^{(t-1)}\)
-
2.
Use random forest to obtain \(y_{i}^{(t-1)} = p_i\) in \(I^{(t-1)}\)
-
3.
Specify variance based on observed data e.g. \(v_i = 0.10\)
-
4.
Calculate \(\alpha _i^{(t-1)}\) and \(\beta _i^{(t-1)}\)
$$\begin{aligned} \alpha _i^{(t-1)} = \frac{p_i^2}{v_i} (1-p_i) \beta _i^{(t-1)} = \frac{p_i}{v_i} (1-p_i)^2 \end{aligned}$$ -
5.
Set prior for probability of forest in area i in image \(\text {I}^{(t-1)}\)
$$\begin{aligned} P_i^{(t-1)} \sim Beta \left( \alpha _i^{(t-1)}, \beta _i^{(t-1)}\right) \end{aligned}$$ -
6.
Form likelihood from image \(\text {I}^{(t-1)}\)
-
7.
Repeat steps 1 to 6 for image \(\text {I}^{(t)}\)
-
8.
Set likelihood for probability of forest in area i in image \(\text {I}^{(t)}\)
$$\begin{aligned} P_i{(t)} \sim Beta(\alpha _i^{(t)}, \beta _i^{(t)}) \end{aligned}$$ -
9.
Compute posterior probability of forest of area i in image \(\text {I}^{(t)}\) as the expectation of the posterior distribution for \(p_i^{(t)}\)
$$\begin{aligned} E(p_i^{(t)})= \frac{\alpha _i^{t} + \alpha _i^{t^{-1}}}{\alpha _i^{t} + \alpha _i^{t^{-1}} + \beta _i^{t} +\beta _i^{t^{-1}}} \end{aligned}$$ -
10.
Use the Beta distribution, \(E(p_i^{(t)})\) as predicted estimate for \(E(p_i^{(t+1)})\)
-
11.
Update \(E(p_i^{(t+2)})\) as the next images become available.
where p is the probability of the observation being forest, as derived from the random forest, and v is some fixed variance. In this case we specify vi based on the variance observed in the testing and training samples across the images, allowing greater variance for \(I^{t-1}\) than for It.
Appendix B: Calculate hyperparameters
From the variance of y
Calculate \(\alpha\)
Use \(\alpha\) to calculate \(\beta\)
Appendix C: Selecting values of variance
In this case study to set variance values we calculated the variance in NDVI and mean NDVI observed in the random samples used for testing and training across the images. We then examined the average variance values to set variance for the images. The average results are below (Table 1):
Based on the sample variance values, we set variance to 0.01 at t and allow a larger value of 0.02 for \(t-1\).
Appendix D: Input probabilities accuracy
The probabilities (p) we use as input variables to calculate \(\alpha\) and \(\beta\) come from a random forest model which makes predictions on each observed image. For example, a random forest is trained on an image at time t to predict probability of forest at time t.
To assess the accuracy of the random forest predicted probabilities, we produced confusion matrices using the true class values of the testing data set and the predicted probabilities performed on the testing data set converted to the class variable using a 0.5 threshold. If the probability of being forest is 0.5 or greater, the probability was converted to forest class and if it was below 0.5 converted to not forest class.
As an example, results for the input random forests for sample 1 at \(t+1\), t and \(t-1\) are as follows (Table 2).
The input random forest for time t + 1 had overall accuracy = 0.985, = Kappa 0.978, = Sensitivity 0.986 and = Specificity 0.985 (Table 3).
The input random forest for time t had overall accuracy = 0.987, Kappa = 0.974, Sensitivity = 0.989 and Specificity = 0.985 (Table 4).
The input random forest for time \(t-1\) had overall accuracy = 0.988, Kappa = 0.975, Sensitivity = 0.990 and Specificity = 0.985.
These results indicate the probability predictions from the input random forests are highly accurate at detecting the true land cover values of forest and not forest in the images at each time point. These are examples of the highly accurate inputs that are then used to calculate the beta distributions used to interpolate missing data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Holloway-Brown, J., Helmstedt, K.J. & Mengersen, K.L. Stochastic spatial random forest (SS-RF) for interpolating probabilities of missing land cover data. J Big Data 7, 55 (2020). https://doi.org/10.1186/s40537-020-00331-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40537-020-00331-8