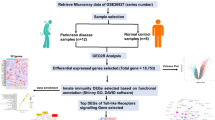
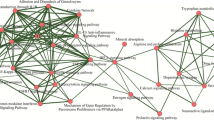
Abstract
Parkinson’s disease (PD) is a progressive neurodegenerative disease affecting the ventral midbrain dopaminergic neurons, resulting in motor defects mainly tremor, rigidity, and bradykinesia along with a wide array of non-motor symptoms. The current study is focused on determining the potential druggable targets of PD by consolidating gene expression profiling and network methodology. Initially, the differentially expressed genes were established from which the central network was constructed by assimilating the interacting partners. Investigating the topological parameters of the network, the genes SYT1, CXCR4, CDC42, KIT, RET, DRD2, NTN1, PRKACB, KDR, NR4A2, SLC18A2, CCK, TH, KCNJ6, and TAC1 were identified as the hub genes and can be explored as potential candidate genes for PD therapeutics. Gene ontology and cluster analysis of the hub genes has provided further insights about the pathophysiology of the disease. Among the hub genes, PRKACB is observed in relatively all the enriched pathways which are modulated by G protein-coupled receptors through protein kinases. Further, we noticed SYT1 as a novel biomarker for PD. Moreover, the regulatory network was constructed with the hub genes as seed nodes with associated transcription factors (TFs) and microRNA (miRNAs). In this analysis, we identified MYC as the major TF and the miRNAs miR-21, miR-155, miR-7, and miR26A1 have a significant role in modulating the hub genes. Briefly, these significant hub genes and their enriched pathways, TFs, and miRNAs have aided in the better understanding of molecular mechanisms underlying PD and its potential core target genes.









Similar content being viewed by others
References
Adams JD Jr, Chang ML, Klaidman L (2001) Parkinson’s disease-redox mechanisms. Curr Med Chem 8:809–814. https://doi.org/10.2174/0929867013372995
Airaksinen MS, Saarma M (2002) The GDNF family: signaling, biological functions and therapeutic value. Nat Rev Neurosci 3:383–394. https://doi.org/10.1038/nrn812
Bader GD, Hogue CW (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4:2. https://doi.org/10.1186/1471-2105-4-2
Ben-Ari Fuchs S, Lieder I, Stelzer G, Mazor Y, Buzhor E, Kaplan S, Bogoch Y, Plaschkes I, Shitrit A, Rappaport N, Kohn A, Edgar R, Shenhav L, Safran M, Lancet D, Guan-Golan Y, Warshawsky D, Shtrichman R (2016) GeneAnalytics: an integrative gene set analysis tool for next generation sequencing, RNAseq and microarray data. OMICS 20:139–151. https://doi.org/10.1089/omi.2015.0168
Chang D, Nalls MA, Hallgrímsdóttir IB, Hunkapiller J, van der Brug M, Cai F et al (2017) A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet 49:1511–1516. https://doi.org/10.1038/ng.3955
Chen Y, Gao Y, Tian Y, Tian DL (2013) PRKACB is downregulated in non-small cell lung cancer and exogenous PRKACB inhibits proliferation and invasion of LTEP-A2 cells. Oncol Lett 5:1803–1808. https://doi.org/10.3892/ol.2013.1294
Chen Y, Lian Y, Ma Y, Wu C, Zheng Y, Xie N (2017) The expression and significance of tyrosine hydroxylase in the brain tissue of Parkinson’s disease rats. Exp Ther Med 14:4813–4816. https://doi.org/10.3892/etm.2017.5124
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY (2014) cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 8:S11. https://doi.org/10.1186/1752-0509-8-S4-S11
Chung S, Leung A, Han BS, Chang MY, Moon JI, Kim CH, Hong S, Pruszak J, Isacson O, Kim KS (2009) Wnt1-lmx1a forms a novel autoregulatory loop and controls midbrain dopaminergic differentiation synergistically with the SHH-FoxA2 pathway.
Decressac M, Volakakis N, Björklund A, Perlmann T (2013) NURR1 in Parkinson disease—from pathogenesis to therapeutic potential. Nat Rev Neurol 9:629–636. https://doi.org/10.1038/nrneurol.2013.209
Dhungel N, Eleuteri S, Li LB, Kramer NJ, Chartron JW, Spencer B, Kosberg K, Fields JA, Stafa K, Adame A, Lashuel H, Frydman J, Shen K, Masliah E, Gitler AD (2015) Parkinson’s disease genes VPS35 and EIF4G1 interact genetically and converge on alpha-synuclein. Neuron 85:76–87. https://doi.org/10.1016/j.neuron.2014.11.027
Dong J, Li S, Mo JL, Cai HB, Le WD (2016) Nurr1-based therapies for Parkinson’s disease. Neurosci Ther 22:351–359. https://doi.org/10.1111/cns.12536
Firestone JA, Smith-Weller T, Franklin G, Swanson P, Longstreth WT, Checkoway H (2005) Pesticides and risk of Parkinson disease: a population-based case-control study. Arch Neurol 62:91–95. https://doi.org/10.1001/archneur.62.1.91
Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) Affy analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20:307–315. https://doi.org/10.1093/bioinformatics/btg405
Goldschneider D, Mehlen P (2010) Dependence receptors: a new paradigm in cell signaling and cancer therapy. Oncogene 29:1865–1882. https://doi.org/10.1038/onc.2010.13
Goldstein DS, Sullivan P, Holmes C, Miller GW, Alter S, Strong R, Mash DC, Kopin IJ, Sharabi Y (2013) Determinants of buildup of the toxic dopamine metabolite DOPAL in Parkinson’s disease. J Neurochem 126:591–603. https://doi.org/10.1111/jnc.12345
He X, Zhang J (2006) Why do hubs tend to be essential in protein networks? PLoS Genet 2:e88. https://doi.org/10.1371/journal.pgen.0020088
Huang DW, Sherman BT, Lempicki RA (2009) Systematics and integrative analysis of large gene sets using using DAVID bioinformatics resources. Nat Protoc 4:44–57. https://doi.org/10.1038/nprot.2008.211
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high-density oligonucleotide array probe level data. Biostatistics 4:249–264. https://doi.org/10.1093/biostatistics/4.2.249
Jha SK, Jha NK, Kar R, Ambasta RK, Kumar P (2015) p38 MAPK and PI3K/AKT signalling cascades in Parkinson’s disease. Int J Mol Cell Med 4:67–86
Kalia LV, Lang AE (2015) Parkinson’s disease. Lancet 386:896–912. https://doi.org/10.1016/S0140-6736(14)61393-3
Karimi-Moghadam A, Charsouei S, Bell B, Jabalameli MR (2018) Parkinson disease from Mendelian forms to genetic susceptibility: new molecular insights into the neurodegeneration process. Cell Mol Neurobiol 38:1153–1178. https://doi.org/10.1007/s10571-018-0587-4
Kennedy TE, Serafini T, de la Torre JR, Tessier-Lavigne M (1994) Netrins are diffusible chemotropic factors for commissural axons in the embryonic spinal cord. Cell 78:425–435. https://doi.org/10.1016/0092-8674(94)90421-9
Lai Wing Sun K, Correia JP, Kennedy TE (2011) Netrins: versatile extracellular cues with diverse functions. Development 138:2153–2169. https://doi.org/10.1242/dev.044529
Lesnick TG, Papapetropoulos S, Mash DC, Ffrench-Mullen J, Shehadeh L, de Andrade M, Henley JR, Rocca WA, Ahlskog JE, Maraganore DM et al (2007) A genomic pathway approach to a complex disease: axon guidance and Parkinson disease. PLoS Genet 3:e98. https://doi.org/10.1371/journal.pgen.0030098
Liu P, Cheng H, Roberts TM, Zhao JJ (2009) Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov 8:627–644. https://doi.org/10.1038/nrd2926
Liu ZP, Wu C, Miao H, Wu H (2015) RegNetwork: an integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database (Oxford):bav095
Mhyre TR, Boyd JT, Hamill RW, Maguire-Zeiss KA (2012) Parkinson’s disease. Subcell Biochem 65:389–455. https://doi.org/10.1007/978-94-007-5416-4_16
Morrison BE (2016) Discovery of nigral dopaminergic neurogenesis in adult mice. Neural Regen Res 11:878–881. https://doi.org/10.4103/1673-5374.184449
Musilli M, Ciotti MT, Pieri M, Martino A, Borrelli S, Dinallo V, Diana G (2016) Therapeutic effects of the Rho GTPase modulator CNF1 in a model of Parkinson’s disease. Neuropharmacology 109:357–365. https://doi.org/10.1016/j.neuropharm.2016.06.016
Pavlidis P, Qin J, Arango V, Mann JJ, Sibille E (2004) Using the gene ontology for microarray data mining: a comparison of methods and application to age effects in human prefrontal cortex. Neurochem Res 29:1213–1222. https://doi.org/10.1023/B:NERE.000
Prakash N, Brodski C, Naserke T, Puelles E, Gogoi R, Hall A, Panhuysen M, Echevarria D, Sussel L, Weisenhorn DM, Martinez S, Arenas E, Simeone A, Wurst W (2006) A Wnt1-regulated genetic network controls the identity and fate of midbrain-dopaminergic progenitors in vivo. Development 133:89e98. https://doi.org/10.1242/dev.02181
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47. https://doi.org/10.1093/nar/gkv007
Satake H (2016) Tachykinin family. In: Yoshio T, Hironori A, Kazuyoshi T (ed) Handbook of hormones: comparative endocrinology for basic and clinical research, 1st edn, AP, San Diego, 72–e9–2. https://doi.org/10.1016/b978-0-12-801028-0.00009-x
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Shimoji M, Pagan F, Healton EB, Mocchetti I (2009) CXCR4 and CXCL12 expression is increased in the nigro-striatal system of Parkinson’s disease. Neurotox Res 3:318–328. https://doi.org/10.1007/s12640-009-9076-3
Smits SM, Ponnio T, Conneely OM, Burbach JP, Smidt MP (2003) Involvement of Nurr1 in specifying the neurotransmitter identity of ventral midbrain dopaminergic neurons. Eur J Neurosci 18:1731–1738. https://doi.org/10.1046/j.1460-9568.2003.02885.x
Steinhoff MS, von Mentzer B, Geppetti P, Pothoulakis C, Bunnett NW (2014) Tachykinins and their receptors: contributions to physiological control and the mechanisms of disease. Physiol Rev 94:265–301. https://doi.org/10.1152/physrev.00031.2013
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C (2017) The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45:D362–D368. https://doi.org/10.1093/nar/gkw937
Tabrez S, Jabir NR, Shakil S et al (2012) A synopsis on the role of tyrosine hydroxylase in Parkinson’s disease. CNS Neurol Disord Drug Targets 11:395–409. https://doi.org/10.2174/187152712800792901
Tang M, Miyamoto Y, Huang EJ (2009) Multiple roles of beta-catenin in controlling the neurogenic niche for midbrain dopamine neurons. Development 136:2027e2038 22
Tu T, Zhang C, Yan H, Luo Y, Kong R, Wen P, Ye CJ, Feng J, Liu F et al (2015) Cd146 acts as a novel receptor for netrin-1 in promoting angiogenesis and vascular development. Cell Res 25:275–287. https://doi.org/10.1038/cr.2015.15
Usiello A, Baik JH, Rougé-Pont F, Picetti R, Dierich A, LeMeur M, Piazza PV, Borrelli E (2000) Distinct functions of the two isoforms of dopamine D2 receptors. Nature 408:199–203. https://doi.org/10.1038/35041572
Vassilatis DK, Hohmann JG, Zeng H, Li F, Ranchalis JE, Mortrud MT, Brown A, Rodriguez SS, Weller JR, Wright AC, Bergmann JE, Gaitanaris GA (2003) The G protein-coupled receptor repertoires of human and mouse. Proc Natl Acad Sci 100:4903–4908. https://doi.org/10.1073/pnas.0230374100
Wallen A, Castro DS, Zetterstrom RH et al (2001) Orphan nuclear receptor Nurr1 is essential for Ret expression in midbrain dopamine neurons and in the brain stem. Mol Cell Neurosci 18:649–663. https://doi.org/10.1006/mcne.2001.1057
Wasterlain CG,Tekchandani L, Liu H, Mazarati AM , Thompson KW ,Suchomelova L , Chen JWY, Niquet J (2009) PEPTIDES | Tachykinins and Epilepsy. Encyclopedia of Basic Epilepsy Research 1117–1123. https://doi.org/b978-012373961-2.00209-5
White CM, Ji S, Cai H, Maudsley S, Martin B (2010) Therapeutic potential of vasoactive intestinal peptide and its receptors in neurological disorders. CNS Neurol Disord Drug Targets 9:661–666
Wolozin B, Behl C (2000) Mechanisms of neurodegenerative disorders: part 1: protein aggregates. Arch Neurol 57:793–796. https://doi.org/10.1001/archneur.57.6.793
Yassen A, Fidel R, Sven-Eric S, Thomas L, Mario A (2008) Computing topological parameters of biological networks. Bioinformatics 24:282–284. https://doi.org/10.1093/bioinformatics/btm554
Yu Q, Liu YZ, Zhu YB, Wang YY, Li Q, Yin DM (2019) Genetic labeling reveals temporal and spatial expression pattern of D2 dopamine receptor in rat forebrain. Brain Struct Funct 224:1035–1049. https://doi.org/10.1007/s00429-018-01824-2
Zhang J, Pho V, Bonasera SJ, Holtzman J, Tang AT, Hellmuth J, Tang S, Janak PH, Tecott LH, Huang EJ (2007) Essential function of HIPK2 in TGFbeta-dependent survival of midbrain dopamine neurons. Nat Neurosci 10:77e86
Acknowledgments
We wish to express our sincere gratitude to VIT University, Vellore, Tamil Nadu, for providing us the facilities for carrying out this research work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
ESM 1
(DOCX 83 kb).
Rights and permissions
About this article
Cite this article
Odumpatta, R., Arumugam, M. Integrative Analysis of Gene Expression and Regulatory Network Interaction Data Reveals the Protein Kinase C Family of Serine/Threonine Receptors as a Significant Druggable Target for Parkinson’s Disease. J Mol Neurosci 71, 466–480 (2021). https://doi.org/10.1007/s12031-020-01669-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-020-01669-7