Abstract
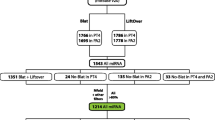
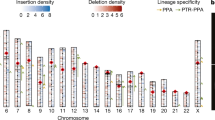
The tripartite motif (TRIM) gene family encodes diverse distinct proteins that play important roles in many biological processes. However, the molecular evolution and phylogenetic relationships of TRIM genes in primates are still elusive. We performed a genomic approach to identify and characterize TRIM genes in human and other six primate genomes. In total, 537 putative functional TRIM genes were identified and TRIM members varied among primates. A neighbor joining (NJ) tree based on the protein sequences of 82 human TRIM genes indicates seven TRIM groups, which is consistent with the results based on the architectural motifs. Many TRIM gene duplication events were identified, indicating a recent expansion of TRIM family in primate lineages. Interestingly, the chimpanzee genome shows the greatest TRIM gene expansion among the primates; however, its congeneric species, bonobo, has the least number of TRIM genes and no duplication event. Moreover, we identified a ~ 200 kb deletion on chromosome 11 of bonobos that results in a loss of cluster3 TRIM genes. The loss of TRIM genes might have occurred within the last 2 mys. Analysis of positive selection recovered 9 previously reported and 21 newly identified positively selected TRIM genes. In particular, most positive selected sites are located in the B30.2 domains. Our results have provided new insight into the evolution of primate TRIM genes and will broaden our understanding on the functions of the TRIM family.



Similar content being viewed by others
Data accessibility
This article does not report new empirical data or software.
References
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L et al (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37(suppl_2):W202–W208
Barr SD, Smiley JR, Bushman FD (2008) The interferon response inhibits HIV particle production by induction of TRIM22. PLoS Pathog 4(2):e1000007
Biris N, Tomashevski A, Bhattacharya A, Diaz-Griffero F, Ivanov DN (2013) Rhesus monkey TRIM5α SPRY domain recognizes multiple epitopes that span several capsid monomers on the surface of the HIV-1 mature viral core. J Mol Biol 425(24):5032–5044
Black LR, Aiken C (2010) TRIM5α disrupts the structure of assembled HIV-1 capsid complexes in vitro. J Virol 84(13):6564–6569
Boesch C, Hohmann G, Marchant L (2003) Behavioural diversity in chimpanzees and bonobos. Cambr Univ Press 109:455–456
Borden KL (1998) RING fingers and B-boxes: zinc-binding protein–protein interaction domains. Biochem Cell Biol 76(2–3):351–358
Boudinot P, Van Der Aa LM, Jouneau L, Du Pasquier L, Pontarotti P, Briolat V et al (2011) Origin and evolution of TRIM proteins: new insights from the complete TRIM repertoire of zebrafish and pufferfish. PLoS ONE 6(7):e22022
Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268(1):78–94
Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F et al (2015) HMMER web server: 2015 update. Nucleic Acids Res 43(W1):W30–W38
Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL et al (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44(D1):D279–D285
Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D et al (2011) Ensembl 2012. Nucleic Acids Res 40(D1):D84–D90
Gokcumen O, Babb PL, Iskow RC, Zhu Q, Shi X, Mills RE et al (2011) Refinement of primate copy number variation hotspots identifies candidate genomic regions evolving under positive selection. Genome Biol 12(5):1–11
Gruber H, Clay ZA (2016) Comparison between bonobos and chimpanzees: a review and update. Evol Anthropol 25:239–252
Gu Z, Cavalcanti A, Chen F-C, Bouman P, Li W-H (2002) Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol Biol Evol 19(3):256–262
Guimarães DS, Gomes MD (2018) Expression, purification, and characterization of the TRIM49 protein. Protein Expres Purif 143:57–61
Han MV, Demuth JP, McGrath CL, Casola C, Hahn MW (2009) Adaptive evolution of young gene duplicates in mammals. Genome Res 19(5):859–867
Han K, Lou DI, Sawyer SL (2011) Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS Genet 7(12):e1002388
Hare B, Wobber V, Wrangham R (2012) The self-domestication hypothesis: evolution of bonobo psychology is due to selection against aggression. Anim Behav 83(3):573–585
Herr AM, Dressel R, Walter L (2009) Different subcellular localisations of TRIM22 suggest species-specific function. Immunogenetics 61(4):271–280
Jiang MXHX, Liao BB et al (2017) Expression profiling of TRIM protein family in THP1-derived macrophages following TLR stimulation. Sci Rep-UK 7:42781–42792
Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C et al (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240
Katoh K, Misawa K, Ki K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30(14):3059–3066
Kieran M, Short TCC (2006) Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J Biol Chem 281:8970–8980
Kovalskyy DB, Ivanov DN (2014) Recognition of the HIV capsid by the TRIM5α restriction factor is mediated by a subset of pre-existing conformations of the TRIM5α SPRY domain. Biochemistry 53(9):1466–1476
Langergraber KE, Prüfer K, Rowney C, Boesch C, Crockford C, Fawcett K et al (2012) Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc Natl Acad Sci 109(39):15716–15721
Letunic I, Doerks T, Bork P (2015) SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43(D1):D257–D260
Li J, Hana K, Xingc J, Kimd H-S, Rogerse J, Ryderf OA et al (2009) Phylogeny of the macaques (Cercopithecidae: Macaca) based on Alu elements. Gene 448(2):242–249
Li M, Zhou XZ, Wang BS (2014) Whole-genome sequencing of the snub-nosed monkey provides insights into folivory and evolutionary history. Nat Genet 46:1303–1310
Maillard PV, Ecco G, Ortiz M, Trono D (2010) The specificity of TRIM5α-mediated restriction is influenced by its coiled-coil domain. J Virol 84(11):5790–5801
Malfavon-Borja R, Sawyer SL, Wu LI, Emerman M, Malik HS (2013) An evolutionary screen highlights canonical and noncanonical candidate antiviral genes within the primate TRIM gene family. Genome Biol Evol 5(11):2141–2154
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY et al (2014) CDD: NCBI's conserved domain database. Nucleic Acids Res 43(D1):D222–D226
McEwan WA, Tam JC, Watkinson RE, Bidgood SR, Mallery DL, James LC (2013) Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat Immunol 14(4):327–336
Meroni G (2012) Genomics and evolution of the TRIM gene family. TRIM/RBCC Proteins, vol 770. Springer, New York, pp 1–9
Oteiza A, Mechti N (2015) Control of FoxO4 activity and cell survival by TRIM22 directs TLR3-stimulated cells toward IFN Type I gene induction or apoptosis. J Interf Cytok Res 35(11):859–874
Pozzi L, Hodgson JA, Burrell AS, Sterner KN, Raaum RL, Disotell TR (2014) Primate phylogenetic relationships and divergence dates inferred from complete mitochondrial genomes. Mol Phylogenet Evol 75:165–183
Prince VE, Pickett FB (2002) Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet 3(11):827–837
Prüfer K, Munch K, Hellmann I, Akagi K, Miller JR, Walenz B et al (2012) The bonobo genome compared with the chimpanzee and human genomes. Nature 486(7404):527–531
Reddy BA, Etkin LD, Freemont PS (1992) A novel zinc finger coiled-coil domain in a family of nuclear proteins. Trends Biochem Sci 17(9):344–345
Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD et al (2006) Global variation in copy number in the human genome. Nature 444(7118):444–454
Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L et al (2001) The tripartite motif family identifies cell compartments. EMBO J 20(9):2140–2151
Sardiello M, Cairo S, Fontanella B, Ballabio A, Meroni G (2008) Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol Biol 8(1):225–247
Sawyer SL, Wu LI, Emerman M, Malik HS (2005) Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA 102(8):2832–2837
Sawyer SL, Wu LI, Akey JM, Emerman M, Malik HS (2006) High-frequency persistence of an impaired allele of the retroviral defense gene TRIM5α in humans. Curr Biol 16(1):95–100
Sawyer SL, Emerman M, Malik HS (2007) Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Pathog 3(12):e197
Sigrist CJ, De Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A et al (2012) New and continuing developments at PROSITE. Nucleic Acids Res 41(D1):D344–D347
Song B, Diaz-Griffero F, Park DH, Rogers T, Stremlau M, Sodroski J (2005) TRIM5α association with cytoplasmic bodies is not required for antiretroviral activity. Virology 343(2):201–211
Suyama M, Torrents D, Bork P (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34(suppl 2):W609–W612
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Tocchini C, Ciosk R (2015) TRIM-NHL proteins in development and disease. Semin Cell Dev Biol 47–48:52–59
Tsai W-W, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S et al (2010) TRIM24 links a non-canonical histone signature to breast cancer. Nature 468(7326):927–932
Uchil PD, Quinlan BD, Chan W-T, Luna JM, Mothes W (2008) TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog 4(2):e16
Versteeg GA, Benke S, García-Sastre A, Rajsbaum R (2014) InTRIMsic immunity: positive and negative regulation of immune signaling by tripartite motif proteins. Cytokine Growth F R 25(5):563–576
Vikram A, Seung-Zin N, Söding J, Lupas AN (2016) The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res 44(W1):W410–W415
Yang Z (1998) Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol 15(5):568–573
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24(8):1586–1591
Yang Z, Nielsen R, Goldman N, Pedersen A-MK (2000) Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155(1):431–449
Zhou X, Sun F, Xu S, Fan G et al (2013) Baiji genomes reveal low genetic variability and new insights into secondary aquatic adaptations. Nat Commun 4:2708
Zhang S, Guo J-T, Wu JZ, Yang G (2013) Identification and characterization of multiple TRIM proteins that inhibit hepatitis b virus transcription. PLoS ONE 8(8):e70001
Zhao G, Ke D, Vu T, Ahn J, Shah VB, Yang R et al (2011) Rhesus TRIM5α disrupts the HIV-1 capsid at the interhexamer interfaces. PLoS Pathog 7(3):e1002009
Acknowledgements
The research was funded by the State Key Program of National Natural Science Foundation of China (Grants no. 31530068), and partial by National Natural Science Foundation of China (no. 31770415) and the Key Research Fund on Sciences and Technologies for Joint Academic Institute and Local Enterprises of Sichuan (2018JZ0008).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declare no conflict of interest.
Additional information
Communicated by Stefan Hohmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Phylogenetic relationships of 537 primate TRIM genes. The unrooted phylogenetic tree was constructed using MEGA 6.0 by Neighbor-Joining method and the bootstrap test was performed with 1,000 iterations. The three groups are indicated as different colors. (TIF 3514 kb)
Rights and permissions
About this article
Cite this article
Qiu, S., Liu, H., Jian, Z. et al. Characterization of the primate TRIM gene family reveals the recent evolution in primates. Mol Genet Genomics 295, 1281–1294 (2020). https://doi.org/10.1007/s00438-020-01698-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-020-01698-2