Abstract
Inferring dynamic system models from observed time course data is very challenging compared to static system identification tasks. Dynamic system models are complicated to infer due to the underlying large search space and high computational cost for simulation and verification. In this research we aim to infer both the structure and parameters of a dynamic system simultaneously by particle swarm optimization (PSO) improved by efficient stratified sampling approaches. More specifically, we enhance PSO with two modern stratified sampling techniques, i.e., Latin hyper cube sampling (LHS) and Latin hyper cube multi dimensional uniformity (LHSMDU). We propose and evaluate two novel swarm-inspired algorithms, LHS-PSO and LHSMDU-PSO, which can be used particularly to learn the model structure and parameters of complex dynamic systems efficiently. The performance of LHS-PSO and LHSMDU-PSO is further compared with the original PSO and genetic algorithm (GA). We chose real-world cancer biological model called Kinetochores to asses the learning performance of LHSMDU-PSO and LHS-PSO in comparison with GA and the original PSO. The experimental results show that LHSMDU-PSO can find promising models with reasonable parameters and structure, and it outperforms LHS-PSO, PSO, and GA in our experiments.
Similar content being viewed by others
1 Introduction
Dynamic system models have been applied to a broad scope of areas, including physics [1], biology [2], chemistry [3], engineering, economics, and medicine [4]. Inferring models of dynamic systems has always been a challenging task. Modelling of dynamic systems are commonly achieved by Ordinary Differential Equations (ODEs), which can capture the overtime evolution of a dynamic system. For an ODE model, a parameter describes the influence rate between variables and a structure describes the interrelation between variables. A set of parameters associated with its structure is considered as the strength of an ODE model [5].
The task of identifying an ODE model from its time series data includes the behaviour modelling of a dynamic system and contrast the simulation outputs with the targeted data. In traditional system identification tasks, normally a model structure is assumed to be given. Therefore, the main aim in these tasks is to identify suitable model parameters that can explain the observed data. However, in many real-world problems, it is very common that the model structure is only partially known or even completely unknown. In such situations, inferring the model, i.e., its structure and parameters, is becoming a more challenging task [6]. This motivates us to explore computational approaches which can automatically learn both the structure and parameters of ODE models simultaneously, as this will be more applicable in many real-world situations when only partial or incomplete knowledge and data are available.
The general form of an ODE can be described by Eq. (1), where \(f_i\) is the relationship among variables, n is the number of observable elements, n and \(X_i\) is the state variable.
For inferring the ODE model of a dynamic system, even the task for simple model with a small number of parameters has been more challenging than expected. This is because practically such models are computationally demanding to simulate. On the other hand, inferring models of complex dynamic systems has even higher computational requirement and needs a feasible point to start with [7]. To achieve our goal, we are using particle swarm optimization (PSO) [8] to infer the structure and parameters of an ODE model. This is because PSO is competent to scaling variables, and it can avoid the premature convergence by effective balancing exploration and exploitation.
PSO has gained much attention in recent years. It is a swarm inspired algorithm first developed by Kennedy and Eberhart [8]. It is a well established meta-heuristic algorithm which provides an adequate solutions to optimize incomplete or imperfect information. In PSO, each particle defines a potential solution and conserves the trajectory of its coordinates in the search space. The trajectory coordinates is related to the solution accomplished known as personal best (pbest). Each particle moves in the search space by updating their coordinates and velocity towards the overall best value known as global best (gbest). Each particle updates its particle coordinates or position after every iteration considering its previous pbest and gbest values as well as its current position and velocity. The particles in a search space are represented by the vector \(X_i=(x_{i1},x_{i2},\ldots ,x_{iD})\), where D is the number of dimensions in the search space. The velocity of a particle in a search space is denoted by \(V_i=(v_{i1},v_{i2},\ldots ,v_{iD})\), as shown in Eq. 2, where \(rand_1\) and \(rand_2\) are random numbers uniformly distributed in the range of [0,1], \(c_1\) and \(c_2\) are positive constants called acceleration, and w is the inertia weight [9].
PSO does not need to adjust too many parameters and in general, it has fast convergence rate, low time, space and design complexity. Moreover, the original version of PSO is for solving continuous problems, but it has been adapted to deal with both discrete and continuous problem search spaces [10, 11].
In this research, we adopt a real biological model and propose to infer both the structure and parameters of its ODE model. We point out that whatever ODE model is under study (biological, chemical or physics), some of its variables may not be realized, which becomes challenging in computation and estimation tasks. We thus suggest the use of a general methodology based on swarm intelligence algorithms that accounts for dynamic systems in a wide range of fields of study. This broadens our ability to cover a large variety of ODE inference problems across different fields.
To our knowledge, the search space of PSO for our problem ( i.e, simultaneously inferring model structure and parameters) is immense even if the number of the variables involved in the search space is small. To overcome this problem, it drove us to search for the best sampling method along with PSO. Therefore, we examine the use of efficient sampling approaches, including Latin Hypercube Sampling (LHS) [12] and Latin Hypercube Sampling Multi Dimensional Uniformity (LHSMDU) [13], as they demonstrates to be effective in many problems with complex search spaces.
To preview other sections of the paper, our study is organised as follows: we discuss the limitations of inferring dynamic system problems along with related work in Sect. 2. In Sect. 3, we discuss the general methodology to address how the model is rifted into structure and parameters, which is fallowed by two main fitness functions to evaluate our model in Sect. 3.1. In Sect. 3.2, we introduce our real cancer biological model. In Sect. 3.3 we presented the proposed algorithms framework. Moreover, In Sect. 3.4, we combine PSO with LHS to show how we can learn or perform the inference of parameters and structure simultaneously. Afterwards, we advanced the original LHS to LHSMDU in Sect. 3.5, to compare the performance difference of LHS-PSO and LHSMDU-PSO. This is followed by using GA in Sect. 3.6, to infer the structure and parameters of the same model for report comparison between proposed algorithms and GA. In Sect. 4, we assess the results and demonstrate the best approach capable of inferring the structure and parameters simultaneously. Finally, we conclude the paper and explore future work in Sect. 5.
2 Literature review
A large number of existing studies in the broader literature have examined the structure and parameters of dynamic systems. First, we will review existing GA approaches as applied to parameters and structure, and this is followed by a review of optimization of ODE. Finally, we will review PSO used for optimizing general function parameters and structure.
Recent research shows that evolutionary algorithms were effective when applied to identify dynamic system models [14,15,16]. In [17], the authors presented an idea to set GA within Genetic Programming (GP). They used GP to optimize the structure of an ODE and GA to optimize its parameters. They have succeeded in inferring an ODE based on observed time series data. However, their experiments were performed by assuming simple equation models ( and thus smaller search space) using the E-Cell Simulation Environment and they wanted to apply the proposed method on real world problems. They claimed the successful inference of structure and parameters but both structure and parameters were inferred separately with different methods of a simple model within small search space. A method of evolutionary approach employed by Yang et al. [18], to concede a gene regulatory network from its time series data. They assumed a simple model and wanted to apply the proposed methodology for inferring a large-scale real model. Previous study points out the problem of assumed simpler model. Mateescu et al. [19] proposed GA for evaluating Cauchy problem and proposed it for real time problems.
Seminal contributions have been made by Darania et al. [20] to carry out a study to estimate the output of differential equations using swarm intelligence. The study falls short of addressing the implementation of methodology on real problems. Lee et al. [21] approximated the outcome of non-linear differential equations by Method of Bilaterally Bounded (MBB). Tan et al. [22] proposed a combined approach to learn a string of solved non-linear differential equations. Mastorakis et al. [23] performed finite element model to assist the fitness functions in search space for identifying an ODE as an optimization problem.
In [24] the author employed PSO and Multi Expression Programming (MEP) to evaluate structural optimization problems. They optimized ODE parameters by PSO and inferred its right-hand side by MEP from a small population in order to retrieve the optimum structure of ODE. They minimized the model complexity by dividing the search space in equal coordinates and proposed the same methodology for real large-scale biochemical network problems. Xu et al. [25] used PSO to track various changes in dynamic functions. They chose Parabolic and Rosenbrock benchmark functions as testing functions because it was easy to control their dynamic change with time. They compared the results of PSO by fixing or changing the global best gbest value for a fixed number of iterations. They suggested that further investigations were needed to assess the performance of PSO with more real world complex models. Similar work was done by Eberhart et al. [26]. They improved the optimization of parabolic function using PSO and kept population size 20 to get speedy results because the system execution was responsive to the number of population and iteration, which describes the optimization of simple functions using PSO with smaller search space is not enough to support the inference of real-world complex models. Previously we have done some work on inferring the model structure and parameters simultaneously for simple models and traditional algorithms [27, 28]. To overcome complex problem and larger search space, we build our work in in Sect. 3 in a way which can infer model structure and parameters of real-world complex models simultaneously.
3 Methodology
For this study, it was of an interest to investigate the application PSO, LHS-PSO, LHSMDU-PSO and GA, to infer both the structure and parameters of an ODE model in parallel. To solve our problem using PSO, the encoding of the search space is the first step. In PSO, each individual location in the search space links one possible solution to its problem. In order to identify the best solution(s), all particles will update their velocity and positions at each iteration. We express the structure as an operation relation between variables. Each particle adjustment point is calculated by a fitness function.
As we mentioned in Sect. 1, the structure of an ODE model outlines the interrelations among variables and the influence rate between variables are represented by the parameters of an ODE model. In general, an ODE model structure comprises Addition, Multiplication, Subtraction and Division (more operations could be added, but this will be explored in future work). The parameters in a model are the coefficients of the variables. A more detailed representation of an ODE is given in Eq. (4), where \(f_1 \sim f_i\) are the state variables, and \(k_1\sim k_i\) are the parameters. Our general model for inferring the structure and parameters of dynamic systems is given in Eq. (4).
To begin with, we are assuming a general model with three different structure item combinations. It includes single variable items i.e. \( k_{1}* x_{1}\), two-variable item i.e. \(k_{1}* x_{2}*x_{3}\) and the item without variable i.e. \(k_{1}\). We can also go further from the scope we set for learning the structure and parameters by assuming more potential terms, but it will be challenging to maintain because the more variable interactions in an ODE, the more complex the design of the solution space. We also argue that many systems may not be very complicated in terms of the number of interactions and variables involved. This is due to the fact that a system needs to maintain robustness while being efficient, and complicated interactions means less robustness and more expensive to function.
We use the addition and subtraction operators to define the interrelation between two items within an equation. Furthermore, we used division or multiplication to define the interrelation between the parameters and variables of an item. Together both parts represent one ODE.
Now, coming towards our objective, we consider a generic setup to define the structure and parameters of an ODE by a particle’s position in the search space. This proceeds in two stages i.e, firstly, the particle dimensions are used to demonstrate each parameter of an ODE in search space. Secondly, we compute all possible structures of an ODE model by a predefined assumption. All assumed structures are then passed into a list \(S_i\) and we use the index value of the list to define the structure. In this way all elements in the list represent the possible structures of a single ODE.
For instance in Table 1, we assume the first five possible structures, each of which is composed of sixteen possible structure item combination, acquired from Eq. 4. In Table 1, each \(S_i\) index value is composed of different combinations of structures. All \(k_i\) (i = 1 to 8) are combined in sequence with structure index \(S_i\) values. The first eight items of the structure \(S_i\) represent the operators i.e, \((+,+,+,+,+,+,+,+)\) a the last eight items of the structure \(S_i\) represents the variable combinations \(x_i\) (where i= 1 to 8) i.e, (\(x_1\), \(x_2\), \(x_3\), \(x_4\), \(x_5\), \(x_6\), \(x_7\), \(x_8\) ).
The main practical problem that confronts us is the sampling distribution of particles in the search space and due to the fact that very little information is know regarding distributing samples in every new iteration. One approach to better normalize the distribution of samples in the search space involves the use of LHS and LHSMDU along with PSO. One of the objectives is to improve the use of LHS and LHSMDU and to relocate the initial particle positions in the search space uniformly. In addition, LHS and LHSMDU provide extra precision and equal coverage for each distribution because they supervise the sampling of each distribution separately. Comparing LHS with Monte Carlo Method, one can say that LHS is outlined accurately to reform the input distribution over sampling in less iterations. LHS stratifies the input probability distributions and stratification partitions the cumulative curve into equal space on the cumulative probability scale. Moreover, LHS can ensure that all particles are distributed in the input search space more evenly, which leads to minor incidents of a sample that is not present in the output distribution of search space.
3.1 The fitness function
The fitness function for each value is expressed as the sum of the squared errors (SSE) as illustrated in Eq. (5), where \(x_i(t_0+k\Delta t)\) is the given target time course data, \(\Delta t\) is the step size, n is the number of state variables, T is the number of the data points, \(t_0\) is the starting time, \(x_i(t_0+k\Delta t)\) and \((k = 0, 1,\ldots , T-1)\) is the time course data attained by measuring the system of ODEs performed by a particle from LHS-PSO.
Model selection turns out to be computationally important for selecting the best fitted model among PSO, LHS-PSO and LHSMDU-PSO. The method of the model selection essentially supports the link between the observations or data and the mathematical model. Some extensive popular methods of rigorous statistical methods have been practiced to analyse model selection criteria. Akaike information criterion (AIC) [29] is one such method which explicitly evaluates the quality of each model. Thus, AIC provides a means for model selection in comparison with other models. In this research, we use the AIC statistical model for comparing the best inferred models and to select the best model. Equation 6 is used to estimate the AIC of a model.
In the above Eq. (6), k is the number of parameters, n is the number of observations in the model and SSE is the Sum of Square Errors. If the number of samples is approximated to be small, i.e, when n/k is less than 40 then a revised model of AIC i.e, AICc is proposed. The best estimation models ranked by AIC is the one with the lowest (most negative) AIC value [29].
3.2 ODE model for biological application
One of the major aim of this study is that we are simulating a real biological cancer tumour model called Kinetochore. Kinetochore is a specific multi-protein set of buildings that performs a vital role in upholding genome consistency [30]. It bridges the gap between micro-tubules and chromosomes during mitosis and operates the spindle meeting checkpoint. A broken kinetochore signifies one possible source for chromosome instability (CIN) and in general CIN plays a role in cancer tumour growth [31]. The obtained model consists of five ODEs. These equations express the rates of change of components d/dt described by Eqs. (7)–(11). Our model is obtained through a collaborative projectFootnote 1 with Dr A. Suarin from University of Dundee and Dr F.Gross from Humboldt University of Berlin, based on their biological experiments and related findings [30]. The model has the following structure as detailed in Eqs. (7)–(11).
In the above Eqs. (7)–(11), the observed variables are \(melt\_p\), \( kln1\_pp1\), \(kln1\_pp2a\), rvsf and \(kln1\_pp2a\_p\). To simplify, we have replaced the variables \(kln1\_pp1\), \(kln1\_pp2a\), \(kln1\_pp2a\_p\), \(melt\_p\) and rvsf with letters T, U, V, W and I respectively. The model variables are presented in a single or combined form of proteins, which are the promoters in human cancer development. In the above mentioned model proteins pp1, pp2a, and p are combined with other main cancer tumour proteins and conjointly they are considered as one variable in our model [32]. Furthermore, variables \(kln1\_pp1 / pp2a /p\) are the number of kinetochore proteins localized in a human cancer cell line [33]. melt is the number of proteins which are strongly associated with cancer and at the same time they serve as the potential targets for drug development against cancer [34]. rvsf is the number of active cancer cells. It is active due to its intensive division that includes replication, transcription, and chromosomal separation [35]. On the other hand, parameter pp1 and pp2a are the numbers of cancer cells to boost cancer in various organs [33], \(sd\_factor\_b\), kinase, and \(darka\_factor\) are the reduced numbers of kln1 and pp2a; \(kd\_kln1\_pp1\) and \(kd\_kln1\_pp2a\) are dissociation constants; \( kp\_melt\) is the phosphorylation constant of protein melt; \(kb\_kln1\_pp1\) and \(kb\_kln1\_pp2a\) are binding constants of pp1 and pp2a respectively and \(kdp\_rvsf\) is the dephosphorylating constant of rvsf.
For instance, in Eq. (12), the values above the over braces describe the structure and the values below the down braces describe the parameters of Eq. (12). Here the \( ``+'' \) sign is representing the structure of variable I and \(``*''\) is representing the interrelation of variable I with parameters pp1, \(sd\_factor\_b\), \(kb\_kln1\_pp1\). Additionally, the sign \(``-''\) is representing the structure of variable T and the sign \(``*''\) represents the interrelation of variable T with parameters \(kd\_kln1\_pp1\). We are taking “*” as a common sign for all variables and parameters presented in an equation. Similarly, we separated all the structures and parameters of our testing model stated in Eqs. (7)–(11), as shown in Table 2.
The time series data form a matrix containing the timestamps and variable values which represent different time steps. In our model the initial concentrations of proteins are as fallows: \(pp1=0.9\), \(sd\_factor\_b = 1\), \(kb\_kln1\_pp1 = 10.24\), \(kd\_kln1\_pp1 = 0.0012\), \(kdp\_pp2a = 0.015\), \(kinase = 1\), \(kp\_pp2a = 0.81\), \(kd\_kln1\_pp2a = 1.64\), \(melt = 0.7\), \(mps1 = 1\), \(kp\_melt = 0.47\), \(pp2a\_p = 0.1\), \(kb\_kln1\_pp2a = 0.12\), \(kdp\_melt = 1591.5\), \(darka\_factor = 1\), \(rvsf\_p = 0.8\), \(kdp\_rvsf = 1410.79\), \(aurb = 1\) and \(kp\_rvsf = 1934.77\). The above mentioned parameter values are initially estimated by the biological modeller (our collaborator) based on experimental data and domain knowledge. Our target model system with parameter values and variables is finally outlined in Eqs. (13)–(17).
For this study, it was of interest to investigate the structure and parameters of the model by considering its performance. As we mentioned earlier, the identified information in an ODE is the given time series data, corresponding parameters and variables. For each ODE the time course data is obtained by means of a function odeintFootnote 2 in Python. It numerically integrates a system of ODEs. The time series data for variables kln1_pp2a_p(V), \(kln1\_pp1 (T), melt\_p\) (W), kln1_pp2a (U),and rvsf (I) are shown in Fig. 1.
3.3 Proposed algorithms framework
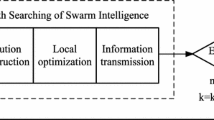
Before we proceed to present the proposed algorithms, we first present the whole framework in Fig. 2 in order to provide a global picture of our research. The whole framework of the proposed algorithms i.e, PSO, LHS-PSO and LHSMDU-PSO is given in Fig. 2.
3.4 The LHS-PSO algorithm
LHS is a commonly used stratified sampling approach due to its effectiveness in initialization of input probability distributions. LHS is designed to achieve a better space-filling performance based on raw samples. It allows the samples to be evenly distributed across the search space.
The basic idea of LHS includes L as an n-dimensional hypercube and \(L^U_i\) and \(L^L_i\) are the upper bound and lower bound of the ith variable \(L_i\), where \(i = 1,2\ldots ,n\). Firstly, to get H samples (solutions), we equally split \(L_i\) range into \(L^U_i\) and \(L^L_i\) into H sub-hypercubes. This way each sub-hypercube will have the same number of dimensions for which the hypercube is initialized. Secondly, it will generate a matrix \(M_{H*n}\) with column and row of n elements. Thirdly, n elements is used as an index value for the selection of sub-hypercubes, and finally we will get one random sampled solution within each sub-hypercubes. For instance, when \(n=2\) and \(H=10\), one potential approach of matrix \(M_{10*2}\) is shown in Table 3.
The outline of \(M_{10*2}\) shows that every dimension in the search space is further divided into 10 sub spaces. Each subspace is indexed by value \([1,2,3,4,\ldots ,10]\). Each sub space is further divided into \(H^2\) sub squares. This way, the possible samples (solutions) (4,4), (2,6), (6,5), (1,2), (3,1), (5,7), (8,8), (6,10), (7,3), (9,2), (10,9) are produced by the LHS method. The number of sub-squares for every single hypercube will be 100 which means that the search space is splited into 100 equal sub-squares.
Figure 3 represents the first three sub-hypercubes, i.e, h0, h1 and h2 of the matrix \(M_{10*2}\) (Table 3). Our proposed LHS-PSO algorithm is shown below.

3.5 The LHSMDU-PSO algorithm
The LHSMDU algorithm enhances the basic idea of LHS. It increases the multidimensional consistency of samples by increasing the distance between realizations. This is accomplished by generating a considerable number of random samples and successively excluding the realizations which are close to each other in the multidimensional space. We assume that a model needs V variables which can be realized W times. The input matrix of this model is composed of V columns and W rows and can be presented by \(V*W\). The sampling matrix composed the range values between \(x^L\) (Lower bound) and \(x^U\) (Upper bound) of the model. Equation 18 describes the sampling matrix of LHSMDU-PSO.
To enhance the multidimensional uniformity, LHSMDU eliminates some realizations that work for any dimension. We used Euclidean distance to eliminate some realization and to calculate the similarity of each realization. The distance \(D_{ij}\) is the dimension of repetition or closeness of realization i and j, which is given by:
In Eq. 19, V is the number of variables with known correlation matrix. We take the two smallest distance values of two nearest neighbours and average them. The average distance is saved for the corresponding realization i and to increment i. The same step is repeated to calculate the average distance of all realizations. The aim of practicing LHSMDU is to validate a larger multivariate space by removing extra realizations from the initial pool and to ease the PSO search for better structures and parameters in the search space. Algorithm.2 defines the LHSMDU-PSO.

3.6 Genetic algorithms
In line with the ideas of inferring the structures and parameters using PSO, LHS-PSO, and LHSMDU-PSO, we also use genetic algorithm to solve the same problem and to make a fair comparison between different algorithms. The execution process of Genetic Algorithm (GA) is not the same as PSO, but its comparison can be discussed. The termination of GA occurs when the fitness level or the given number of generations has been attained. Notably, we exemplify the model in terms of Genome, Chromosome and Gene in Eq. (20). To make one complete genome, firstly we consider a single parameter \(T*0.0012\) as a gene. Secondly we merge all genes to shape a single chromosome and finally we merge all chromosomes to shape one complete genome. Algorithm. (3) outlines GA.

4 Experimental results
We applied PSO, LHS-PSO, LHSMDU-PSO and GA to assess the effectiveness of these algorithms. We performed the experiments of the proposed algorithms on the ODEs of the kinetochore model. We developed the elemental PSO first and then enhanced it with LHS and LHSMDU, later we compared the best algorithm among PSO, LHS-PSO and LHSMDU-PSO with GA for better comparison. All experiments were conducted on a computer with an Intel Core i5 processor (3.20 GHz), 8 GB of RAM.
Considerable attention was given to tuning the parameters of PSO and GA. We selected the best performing parameters for both algorithms during the optimization process. We ran a number of trials based experiments beforehand in order to tune the best performing parameters for solving the ODEs of dynamic systems. In addition, as we discussed, dynamic systems are very complex and it is very challenging to learn the structure and parameter simultaneously. One way to help overcome this challenge is to keep the population size and iteration high. It will help to enhance the exploration capability in the search space to discover better particles. Parameters set for the GA and PSO algorithms are the same in all experiments. In PSO, the number of iterations is 1,000, the size of population 1,000, \(\omega \) is 0.6, \(c_1\) and \(c_2\) are 0.8. In GA, the population size is 1000, crossover probability is 0.85, mutation probability is 0.3 and the number of generations is 500. We will further compare the best algorithm among PSO, LHS-PSO and LHSMDU-PSO with GA.
The following observations were made by applying PSO, LHS-PSO and LHSMDU-PSO: we acquired the best fitness values by means of sum of square errors. The corresponding ODE models for PSO, LHS-PSO and LHSMDU-PSO are described in Eqs. (21)–(25), (26)–(30) and (31)–(35) respectively.
1. The Resultant ODE Model found by PSO
2. The Resultant ODE Model found by LHS-PSO
3. The Resultant ODE Model found by LHSMDU-PSO
For each algorithm we conducted 20 trials of experiments. This allowed us to compare LHSMDU-PSO with LHS-PSO and PSO. The fitness values shown in Table 4 and Fig. 4 indicate a clear significant difference. They demonstrate that LHSMDU-PSO algorithm has a better ability to search for the parameters and structure of the system of ODEs by using known time course data. Furthermore, if none of the algorithms found the right structure, this is still valuable as the problem is complicated in nature. In addition, the alternative models found by our algorithms provide additional insight and guidance. Finally, we noticed that LHSMDU-PSO can find models which are closer to the measured data than other algorithms, and this indicates the performance of LHSMDU-PSO.
The time course data generated for PSO, LHS-PSO and LHSMDU-PSO in Fig. 5 shows that the corresponding ODE model we constructed through LHSMDU-PSO matches the initial time course data more closely.
Furthermore, to assess the significant result group among PSO, LHS-PSO and LHSMDU-PSO, we used Kruskal–Wallis H statistical test [36], which shows the significant group between two or more independent groups. We carried out an experiment to see if PSO, LHS-PSO or LHSMDU-PSO are better on the basis of their fitness functions. Table 5 shows the Kruskal–Wallis Hypothesis test summary for the given PSO, LHS-PSO and LHSMDU-PSO.
In the above Table 5, the p value is: \(p<0.001\), which shows a very strong evidence of a difference between at least one pair of groups. Since the p value is less than .001, we did post hoc analysis to see which group was significantly different from the other. Table 6, shows the group wise comparison of PSO, LHS-PSO and LHSMDU-PSO.
From post hoc analysis in Table 6, we can say that LHSMDU-PSO is significantly different from PSO among other comparisons. The mean rank of PSO, LHS-PSO and LHSMDU-PSO is 49.85, 30.05 and 11.60 respectively which indicates that LHSMDU-PSO performs the best in comparison with PSO and LHS-PSO.
We earlier mentioned and discussed the importance of AIC in model selection. Since n/k is greater than 40, so we calculated AIC instead of AICc for every possible combination of variables. One of the key factors of calculating AIC is to show AIC scores and their models. The result of AIC is shown in Table 7.
By comparing the scores of PSO, LHS-PSO and LHSMDU-PSO, we can see that LHSMDU-PSO is the best approximation model in comparison with the best models of PSO and LHS-PSO. Furthermore, we can also find the best approximation model by calculating the difference \(\Delta i\) between the best model of AIC with each other model.
All the above statistical and practical results indicate that LHSMDU-PSO is promising in order to find a better structure and parameters of target ODE model. Therefore, we will further compare it with GA in more details. The corresponding ODE models for GA is described in Equations. (36)-(40).
1. The Resultant ODE Model of GA
GA provides a considerable insight in inferring parameters and structure of an ODE. This investigations so far are too small for a fair comparison, so we tried to analyse which algorithm quickly infer the better structure and parameters by giving them the same amount of time. A time frame of 600 s were given to GA and LHSMDU and they were started at the same time. Eight trials were executed, neither GA or LHSMDU-PSO infer the structure and parameters in the given time frame. Furthermore, we did 10 more trials and calculated the time in which GA and LHSMDU-PSO trials are ended upon the completion of its evolution and iteration number respectively. The time taken to infer both the structure and parameters from both GA and LHSMDU-PSO are presented in Fig. 6 and Table 8.
We calculated the Average Time, Maximum Time and Standard Deviation of PSO, LHS-PSO, LHSMDU-PSO and GA. Among GA and LHSMDU-PSO, it highlights that GA requires 30.78 % more time on average than LHSMDU-PSO. In the first step, the time course data generated by GA in Figs. 5 and 6, underlines that the corresponding ODE model constructed by GA is not closely matching the initial time course data as compared to LHSMDU-PSO. This result has further strengthened our confidence in LHSMDU-PSO and demonstrates its clear advantage over GA.
The results of the experiment found clear support for the LHSMDU-PSO. We have verified that using the sampling technique of LHSMDU can produce significantly better results. The key leverage of LHSMDU-PSO among other sampling technique is that it spreads all the particles in the search space evenly, and by calculating the Euclidean distance among each other, the smallest average distance to its two nearest neighbors is eliminated. The data points corresponding to the uniformly sampled points are picked up.
5 Conclusions and future work
This is an important finding in understanding the inference of the structure and parameters of dynamic systems. To the best of our knowledge, this is the first report that proposed the identification of system of ODEs from observed time course data by means of swarm-inspired approaches enhanced by stratified sampling techniques. The use of LHS and LHSMDU along with PSO improves the original PSO. It assigns the particles initial positions rather than initializing them randomly. The findings provide a potential mechanism to achieve a system of ODEs which is closely matching to the observed time series data. We have shown that the proposed algorithm performs better than standard PSO and GA. The evidence from this study implies that LHSMDU-PSO infers the structure and parameters of ODE models more efficiently. This is ascribed to the effectiveness of PSO, which is further enhanced by effective sampling techniques (LHSMDU). This also shows the promising application potential of LHSMDU-PSO in solving other problems with hybrid discrete and continuous search spaces in a broader range of fields.
In our future research, we would like to further investigate the proposed algorithm and to extended it to infer more complex models with more mathematical operators, such as the exponential operator. Furthermore, apart from i more complex models and exponential operators, future research should look to extend our work to infer semi-quantitative models [37].
Notes
scipy.integrate.odeint.
References
Melby P, Weber N, Hübler A (2005) Dynamics of self-adjusting systems with noise. Chaos Interdiscip J Nonlinear Sci 15(3):033902
Gintautas V, Foster G, Hübler AW (2008) Resonant forcing of chaotic dynamics. J Stat Phys 130(3):617
Jackson T, Radunskaya A (2015) Appl Dyn Syst Biol Med, vol 158. Springer, Berlin
Gandolfo G (1971) Economic dynamics: methods and models, vol 16. Elsevier, New York
Boyen X, Friedman N, Koller D (1999) Discovering the hidden structure of complex dynamic systems. In: Proceedings of the fifteenth conference on uncertainty in artificial intelligence. Morgan Kaufmann Publishers Inc., Burlington, pp 91–100
Heinonen M, Yildiz C, Mannerström H, Intosalmi J, Lähdesmäki H (2018) Learning unknown ODE models with Gaussian processes. arXiv preprint arXiv:1803.04303
Cleghorn CW, Engelbrecht AP (2017) Proceedings of the genetic and evolutionary computation conference. ACM, pp 12–18
Eberhart J, Kennedy J (1995) Particle swarm optimization. In: Proceedings of the IEEE international conference on neural networks, vol 4 (Citeseer, 1995), pp 1942–1948
Zhang L, Yu H, Hu S (2003) A new approach to improve particle swarm optimization. In: Genetic and evolutionary computation conference. Springer, Berlin, pp 134–139
Pang W, Wang Kp, Zhou Cg, Dong Lj (2004) Fuzzy discrete particle swarm optimization for solving traveling salesman problem. In: The fourth international conference on computer and information technology. CIT’04. IEEE, pp 796–800
Huang L, Wang K, Zhou C, Pang W, Dong L, Peng L (2003) Particle swarm optimization for traveling salesman problems. Acta Scientiarium Naturalium Universitatis Jilinensis 4:12
Helton JC, Davis FJ (2003) Latin hypercube sampling and the propagation of uncertainty in analyses of complex systems. Reliab Eng Syst Saf 81(1):23
Deutsch JL, Deutsch CV (2012) Latin hypercube sampling with multidimensional uniformity. J Stat Plan Inference 142(3):763
Karr CL, Wilson E (2003) A self-tuning evolutionary algorithm applied to an inverse partial differential equation. Appl Intell 19(3):147
Reich C (2000) Simulation of imprecise ordinary differential equations using evolutionary algorithms. In: Proceedings of the 2000 ACM symposium on Applied computing-Volume 1. ACM, pp 428–432
Cao H, Kang L, Chen Y, Yu J (2000) Evolutionary modeling of systems of ordinary differential equations with genetic programming. Genetic Program Evol Mach 1(4):309
Iba H (2008) Inference of differential equation models by genetic programming. Inf Sci 178(23):4453
Yang B, Chen Y, Meng Q (2009) Inference of differential equation models by multi expression programming for gene regulatory networks. In: International conference on intelligent computing. Springer, pp 974–983
Mateescu GD et al (2006) On the application of genetic algorithms to differential equations. Rom J Econ Forecast 7(2):5–9
Babaei M (2013) A general approach to approximate solutions of nonlinear differential equations using particle swarm optimization. Appl Soft Comput 13(7):3354
Lee ZY (2006) Method of bilaterally bounded to solution Blasius equation using particle swarm optimization. Appl Math Comput 179(2):779
Liao S, Tan Y (2007) A general approach to obtain series solutions of nonlinear differential equations. Stud Appl Math 119(4):297
Mastorakis NE (2006) Unstable ordinary differential equations: solution via genetic algorithms and the method of Nelder-Mead. WSEAS Trans Math 5(12):1276
Gandomi AH, Yang XS, Alavi AH (2013) Cuckoo search algorithm: a metaheuristic approach to solve structural optimization problems. Eng Comput 29(1):17
Hu X, Eberhart RC (2002) Evolutionary computation, 2002. CEC’02. Proceedings of the 2002 Congress on, vol 2. IEEE, pp 1666–1670
Eberhart RC, Shi Y (2001) Evolutionary computation, 2001. Proceedings of the 2001 congress on, vol 1. IEEE, pp 94–100
Usman M, Awad A, Pang W, Coghill GM (2019) Proceedings of the genetic and evolutionary computation conference companion. ACM, pp 101–102
Tian X, Pang W, Wang Y, Guo K, Zhou Y (2019) LatinPSO: an algorithm for simultaneously inferring structure and parameters of ordinary differential equations models. Biosystems 182:8–16
Symonds MR, Moussalli A (2011) A brief guide to model selection, multimodel inference and model averaging in behavioural ecology using Akaike’s information criterion. Behav Ecol Sociobiol 65(1):13
Nijenhuis W, Vallardi G, Teixeira A, Kops GJ, Saurin AT (2014) Negative feedback at kinetochores underlies a responsive spindle checkpoint signal. Nat Cell Biol 16(12):1257
Yuen KW, Montpetit B, Hieter P (2005) The kinetochore and cancer: what’s the connection? Curr Opinion Cell Biol 17(6):576
Fujiki H, Suganuma M (2009) Marine toxins as research tools. Springer, Berlin, pp 221–254
Takimoto M, Huret JL (2017) Atlas of genetics and cytogenetics in oncology and haematology. Nucl Acids Res 28(1):349–351
Singh V, Ram M, Kumar R, Prasad R, Roy BK, Singh KK (2017) Phosphorylation: implications in cancer. Protein J 36(1):1
Tadic M, Cuspidi C, Hering D, Venneri L, Danylenko O (2017) The influence of chemotherapy on the right ventricle: did we forget something? Clin Cardiol 40(7):437
Guo S, Zhong S, Zhang A (2013) Privacy-preserving Kruskal–Wallis test. Computer Methods Programs Biomed 112(1):135
Pang W, Coghill GM (2015) Qualitative, semi-quantitative, and quantitative simulation of the osmoregulation system in yeast. BioSystems 131:40
Acknowledgements
We would like to thank Dr. F. Gross from Humboltd University for providing us with the original ODE models for Kinetochores.
Funding
This research is funded by Elphinstone Scholarship provided by University of Aberdeen, Scotland, UK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Muhammad Usman has received research grants from University of Aberdeen, Scotland, UK. Muhammad Usman, Wei Pang and George M.Coghill declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Usman, M., Pang, W. & Coghill, G.M. Inferring structure and parameters of dynamic system models simultaneously using swarm intelligence approaches. Memetic Comp. 12, 267–282 (2020). https://doi.org/10.1007/s12293-020-00306-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12293-020-00306-5