Abstract
Acute respiratory distress syndrome (ARDS) is a kind of comprehensive disease with excessive inflammation and high clinical mortality. Multiple immune cells are involved in the ARDS process. Amongst these populations, lung-resident alveolar macrophages (AMs) are known to participate in the regulation of ARDS. GPR84, a metabolite-sensing GPCR sensing medium-chain fatty acids (MCFAs), is highly expressed in LPS-challenged macrophages and considered as a pro-inflammatory receptor. In this study, it was hypothesized that Gpr84 may be involved in pulmonary homeostasis via its regulatory effect on the switch of AM status. In LPS-induced ALI mouse model, we identified the internal LPS-induced switch of AMs from CD11blo to more inflamed CD11bhi status, which is deeply related to the exacerbated imbalance of homeostasis in the lung injury process. Gpr84 was highly expressed in ALI lung tissues and involved in cytokine release, phagocytosis and status switch of AMs through positive regulatory crosstalk with TLR4-related pathways via CD14 and LBP, which relied on Akt, Erk1/2, and STAT3. If conserved in humans, GPR84 may represent a potential therapeutic target for ARDS.
Similar content being viewed by others
Introduction
Acute respiratory distress syndrome (ARDS), formerly also referred as acute lung injury (ALI), is a non-cardiogenic pulmonary oedma1,2 complicated by systemic inflammation, including excessive release of inflammatory mediators TNF, IL-6, IL-12, and immune cell infiltration.3,4,5,6 Recent epidemiology data have revealed direct and indirect risk factors of ARDS, amongst which pneumonia, the aspiration of gastric contents, and sepsis cause more than 85% of cases.2
Balanced pulmonary homeostasis is crucial for the physiological functions of lung, which becomes disrupted during ARDS.7,8,9 Due to the existence of the lung epithelial barrier, the pulmonary immune homeostasis is relatively self-governed by a complex regulatory network, including but not limited to resident alveolar macrophages (AMs),10,11,12,13 neutrophils,14,15,16,17 T lymphocytes18,19,20, B lymphocytes,21 NK cells,22 and alveolar epithelial cells.23,24,25 Amongst these cell populations, AMs and recruited monocytes/macrophages play pivotal roles in the pathogenesis of ARDS. In ARDS, AMs are source of IL-1α,26 IL-1β,27 TNF,28 CXCL1,29 CXCL2,27 and type I IFNs.30 AMs also contribute to the neutrophil recruitment,31 the regulation of Treg,32,33 and the phagocytosis of surfactants.34,35,36 Nevertheless, the regulatory mechanisms of AMs in ARDS are still unclear.
The characteristics of myeloid-derived monocytes/macrophages (MO/Mφ) differ to AMs,37,38,39 which are embryonic-origin lung-resident macrophages that mainly self-renew during whole lifespan.38,40,41 Recent studies have revealed that metabolic reprogramming regulated the switch between the pro- and anti-inflammatory status of macrophages, which may differ amongst myeloid-derived and tissue-resident macrophages. In myeloid-derived MO/Mφ, glycolysis is the main metabolic mechanism in classical macrophage activation, whilst fatty acid oxidation (FAO) and OXPHOS used to be considered as dominate metabolic pathways in the alternative macrophage activation.42,43,44,45 However, recent studies showed that both FAO and OXPHOS were dispensable in alternative macrophage activation.46 AMs are committed to FAO at steady state,47,48 which differs from myeloid-derived MO/Mφ and lung interstitial macrophages.47 In the light of above, varied relationships of immunometabolism and macrophage activation are worthy of study.
Free fatty acids (FFAs) are essential sources for cellular functions.49 FFAs commonly originate from the decomposition of triglycerides50 and the catabolites of symbiotic bacteria,51 whilst lipopolysaccharide (LPS) stimulation may enhance the uptake of FFAs in macrophages.52 Cellular uptake of exogenous FFAs may contribute to de novo fatty acid synthesis,53,54 survival,55 and immune responses. Free fatty acid receptors (FFARs) are a group of metabolite-sensing GPCRs,56,57 including GPR40, GPR41, GPR43, GPR84, GPR119, and GPR120, that use FFAs as ligands and are expressed differentially in immune cells and involved in the regulation of inflammation. GPR41 and GPR43 can promote neutrophil recruitment to infected tissues,58 whilst GPR40 and GPR120 repress tissue inflammation.59,60 Amongst FFARs, GPR84 plays a potential role in pro-inflammatory processes. GPR84, also termed GPCR4 or EX33, was first discovered in neutrophils,61 and can be activated by medium-chain fatty acids (MCFAs, C9–C14) in relatively high doses.62 The expression of Gpr84 was upregulated in microglia63 and 3T3-L1 adipocytes64 in response to LPS or TNF stimulation in mice, respectively. Activation with agonists or MCFAs to GPR84 could enhance the mRNA levels of pro-inflammatory mediators, including Il12b,62 Tnf,65 and IL8,65 whilst Gpr84 deficiency reduced the mRNA levels of Il6, Tnf, and Ccl2.66 GPR84 downstream pathways may refer to Gi/o,62 Akt, Erk1/2, and NF-κB.67 However, the clear roles of GPR84 during inflammation require further elucidation.
In this study, LPS-induced ALI mouse model was established through intratracheal instillation of LPS to mimic and clarify the regulatory role of GPR84 in AMs during the inflammatory process in ARDS. Consistent with the perspective of restricted alveolar niches,68 our results indicated that the absolute CD45.2+F4/80+ cells in BALF remained relatively constant during ALI process, the majority of which are Ly-6G−CD45.2+F4/80+CD11a+Siglec-F+Ly-6CmidCD11c+CCR2loCX3CR1lo-midCD14lo-midCD326lo-midCD11blo-hi AMs. AMs interacted with LPS and switched from CD11blo to CD11bhi status with enhanced inflammatory cytokine release and phagocytic capacity, formerly considered as recruited monocytes or M1-like AMs. Gpr84 deficiency significantly alleviated tissue necrosis, downregulated neutrophil infiltration, and the release of pro-inflammatory cytokines. These regulatory effects of Gpr84 may be attributed to the involvement in the switch of AMs status through the crosstalk with TLR4-related pathways via Akt/Erk1/2-STAT3-CD14/LBP. This study provides insights into the role of GPR84 as a potential therapeutic target for ARDS.
Results
Dynamic switch of AM status occurs in LPS-induced ALI mouse model
Different cell populations (Gating strategies see Fig. 1f, Supplementary Fig. 1a, c) in pulmonary microenvironment were preliminarily investigated in the progress of LPS-induced ALI mouse model. Ly-6G−CD45.2+F4/80+CD11a+Siglec-F+Ly-6CmidCD11c+CCR2loCX3CR1lo-midCD14loCD326lo-midCD11blo AMs dominate the alveolar niches at steady state, whilst time-dependent Ly-6G+CD11b+ neutrophil infiltration happened during ALI process (Fig. 1a). Absolute amounts of CD45.2+F4/80+ cells in alveolar niches remained relatively constant (Fig. 1a), with slightly decrease of AMs and subsequent increase of recruited Ly-6G−CD45.2+F4/80+CD11a+Siglec-F−CD11c−CCR2mid-hiCD14hiCD11b+Ly-6Cmid-hi MOs, which partly verified the perspective of restricted alveolar niches.68 The recruited Siglec-F−CCR2hiCD11b+Ly-6Chi MOs were mainly discovered in lung parenchyma, whilst AMs still dominate the CD45.2+F4/80+ cells in alveoli niches (Fig. 1a, Supplementary Fig. 1d).
a Change of different BALF cell populations during the ALI process; b Trends of IL-6, TNF, IL-12 and CCL2 release in ALI; c Comparison of different AM markers between sham-AMs and ALI-AMs (four biological repeats for each time point, two technical repeats); d Change of different BALF cell populations with ALI or sham treatment that received PBS- or clodronate-liposomes 24 h before ALI; e Influence of clodronate-induced AM depletion on ALI-related cytokine release; f Unbiased clusters of lung parenchyma cells based on gating strategy; g LPS-bind capacity of AMs in vivo implied by the MFI of LPS-FITC. Data of a, b are shown as means ± SEM (four biological repeats, two technical repeats), asterisk symbol indicates (*) compared with sham group; data of d, e are shown as means ± SEM (four and eight biological repeats for sham and ALI group, respectively, two technical repeats each); *0.01 < P < 0.05; **P < 0.01; ***P < 0.001.
Further exploration of AMs from mice with sham treatment or LPS challenge indicated a time-dependent switch to CD11bhi status (Fig. 1a) with several changes of AM markers (Fig. 1c). ALI led to AMs with increased FSC, partially indicating morphological changes as well as enhanced phagocytic potential of ALI-CD11bhi AMs. With time-dependent increasing expression of CD11b on AMs in ALI mice, elevated level of F4/80 and slightly upregulated level of CD11c were also found, possibly implying a more activated status. In contrast, CD11bhi AMs showed lower levels of Siglec-F and CD326, which were usually considered as cardinal markers of AMs at steady state. The switch of AM status coincided with ALI-related cytokine release in BALF (Fig. 1b), possibly supporting the correlation between dynamic switch of AM status and the ALI process, whilst first peaks of TNF and IL-6 occurred at 6 h implied an acute inflammatory response.
AMs play a vital role in ALI and CD11bhi AMs act as a more inflamed status
The regulatory functions of AMs at different status in ALI require further elucidation. Temporary depletion of AMs with clodronate liposomes was applied and resulted in significantly decreasing of neutrophil infiltration and ALI-related cytokine release (Fig. 1e), which indicated that AMs were involved in the regulation of neutrophil invasion and inflammatory cytokine release during ALI.
Considering the complexity of pulmonary immune homeostasis, FITC-conjugated LPS was used to explore cell populations that directly interact with LPS in vivo. AMs displayed the strongest FITC signal which indicated superior capacity to interact LPS (Fig. 1g), whilst other cell populations showed negative or weak signals (Supplementary Fig. 2a). Thus, AMs were the main cell populations that directly bind to LPS and initiate ALI process. The biased LPS–AM interaction may be a result of unique alveolar structure, leading to inefficient transport of LPS into the lung parenchyma. Based on above, we have reason to believe that AMs were key cells to initiate further immune responses under inhaled challenge, such as the inhaled pneumonia caused by Gram-negative bacteria.
AMs were generally considered as immunosuppressive at steady state. However, the actual phenotypes and functions of AMs in ALI remained controversial. In this study, Sham-CD11blo AMs, ALI-CD11bint, and ALI-CD11bhi AMs were sorted (Fig. 2a) and analyzed. ALI-related cytokines were highly expressed in CD11bhi AMs (Fig. 2b) which may represent a more inflamed status. Upregulated expressions of macrophage activation markers (Fig. 2b) in CD11bhi AMs hinted that the switched status of AMs in ALI cannot be classified as pro- or anti-inflammatory exclusively, but represented an intermediate status with combined functions. In addition, CD11bhi AMs showed higher levels of TNF, IL-6, IL-12 secretion, and enhanced phagocytic capacity amongst the AM status (Fig. 2c, d), which reinforced the role of CD11bhi AMs as a more pronounced inflamed status.
a Sorting strategy of AMs with different status in mice with sham or ALI treatment; b Expressions of macrophage activation markers and ALI-related cytokines in AMs with different status in vivo; c Secretion of TNF, IL-6, and IL-12 in AMs with different status in vivo; d Phagocytic capacity of AMs with different status in vivo; e Expressions of FFARs in lung tissue, BALF cells, and parenchyma cells from mice with sham or ALI treatment; f Expressions of FFARs in neutrophils and AMs in vivo; g Expressions of FFARs in AMs with different status in vivo. Data of b, g are shown as means ± SEM (n = 3), hash symbol indicates (#) compared with sham-CD11blo AMs, asterisk symbol indicates (*) compared with ALI-CD11bmid AMs; Data of e, f are shown as means ± SEM (n = 3), asterisk symbol indicates (*) compared with sham group, *0.01 < P < 0.05; **P < 0.01; ***P < 0.001.
Gpr84 was involved in the dynamic switch of AM status in ALI
Our previous gene microarray results showed Gpr84 was highly expressed in lung tissues from LPS-induced ALI mice (data not shown). Amongst the metabolite-sensing FFARs, Gpr84 was uniquely and highly expressed in lung tissues as well as in BALF and parenchyma cells from ALI mice (Fig. 2e). To specify the distribution of Gpr84 expression, different immune cells were sorted and assessed. AMs and neutrophils, two main effector cells in ALI, had higher Gpr84 expression than other FFARs (Fig. 2e, f). Other immune cells, including CD19−CD3+NK1.1− T lymphocytes, CD3−CD19+ B lymphocytes and CD19−CD3−NK1.1+ NK cells, showed a similar variation to all FFARs (Supplementary Fig. 2b). Furthermore, we found that the expression of Gpr84 was the highest in CD11bhi AMs amongst AM status (Fig. 2g). Based on these results, we assumed that GPR84 is likely to be involved in ALI process.
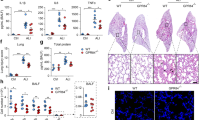
To reveal the roles of Gpr84 in ALI, Gpr84 deficient mice were constructed via CRISPR/Cas9 approaches. Both male and female Gpr84 deficient mice showed a little weight loss (Supplementary Fig. 3a). Various histological analysis consistently indicated that Gpr84 deficiency alleviated the impairment of structural remodeling, MPO activity, lung permeability, cell invasion, and cytokine release in ALI mice (Fig. 3a–d). Although Gpr84 deficiency seemed not to affect the absolute CD45.2+F4/80+ cells in BALF, the progress of the AM switch during ALI process was delayed (Fig. 3e). The influence of Gpr84 on status switch was further verified through chimeric BMT (Fig. 3f, Supplementary Fig. 3b, c). In BMT, chimeric BM cells with different ratio (the ratio was calculated as injected number of CD45.1+WT BM cells to that of CD45.2+Gpr84−/− BM cells) were transplanted to explore the difference of WT and Gpr84−/− AMs in the same individual system, whilst WT to WT group and Gpr84−/− to Gpr84−/− group were used as controls. In addition to the validation of delayed status switch of AMs, the influence of Gpr84 knockout on chemotaxis of different immune cells were investigated (Supplementary Fig. 3d). The regulatory effect on the switch of AM status may be a key mechanism of Gpr84 in ALI, since the role of AM switch had been described above.
a Histology of lung structural remodeling in WT and Gpr84−/− mice shown with HE staining; b MPO activity in WT and Gpr84−/− mice detected with IHC; c Lung permeability analysis of WT and Gpr84−/− mice with Evans blue injection through caudal vein; d ALI-related cytokine release in BALF from WT and Gpr84−/− mice; e Neutrophil and monocyte infiltration in WT and Gpr84−/− mice; f Comparison of cell infiltration and status switch in BM-transplanted mice with different chimeric ratio. Data of c–f are shown as means ± SEM (four and eight biological repeats for sham and ALI group, respectively, two technical repeats each), *0.01 < P < 0.05; **P < 0.01; ***P < 0.001.
Gpr84 regulates LPS-induced inflammation via CD14 and LBP
Previous studies regarded GPR84 as a pro-inflammatory receptor in immune cells.65 However, the exact relationships and underlying mechanisms within Gpr84 and inflammatory reactions require elucidation. To explore the preliminarily functions of Gpr84 on AMs, MH-S cell line were employed. Similar to RAW264.7 (data not shown), Gpr84 was highly expressed in LPS-stimulated MH-S (Fig. 4a). Combined stimulation with the GPR84 agonist diindolyl-methane enhanced LPS-induced upregulated expressions of several ALI-related cytokines in MH-S, including Il6, Il12b, and Cxcl2 (Fig. 4b). Knockdown of Gpr84 with siRNA (Fig. 4c) prior to LPS stimulation significantly downregulated the mRNA level of LPS-induced cytokines (Fig. 4d).
a Gpr84 expression in alveolar macrophage cell line MH-S under LPS challenge. b Expressions of ALI-related cytokines in MH-S with combined stimulations of LPS and GPR84 agonists; c Efficiency of Gpr84 knockdown in MH-S by RNA interference; d Expressions of ALI-related cytokines in MH-S after Gpr84 knockdown; e–g Expressions of ALI-related cytokines in Gpr84+/+ and Gpr84−/− AMs, BMDMs, and PEMs; h Phagocytosis of latex beads in Gpr84+/+ and Gpr84−/− AMs; i LPS-induced CD11b switch in Gpr84+/+ and Gpr84−/− AMs. Data of a–g are shown as means ± SEM (four biological repeats, three technical repeats), asterisk symbol indicates (*) compared with DMSO/NC/Gpr84+/+ cells under LPS challenge; data of h, i are shown as means ± SEM (four biological repeats, two technical repeats each), *0.01 < P < 0.05; **P < 0.01; ***P < 0.001.
Primary AMs were isolated from Gpr84 deficient mice to further study the role of Gpr84. AM markers showed minor difference between WT and Gpr84−/− AMs in vitro (Supplementary Fig. 4a). Tracing of chimeric transplanted CD45.1+WT and CD45.2+Gpr84−/− bone-marrow cells showed lower level of several AM markers throughout AM repopulation in vivo (Supplementary Fig. 5). WT and Gpr84−/− AMs, BMDMs, and PEMs, from different sources and tissues in mice, were isolated and stimulated with LPS to investigate the functions of Gpr84 in macrophages. The enhanced levels of LPS-induced cytokines decreased in Gpr84−/− macrophages compared to WT (Fig. 4e–g). Gpr84 deficiency was also found to weaken the AM phagocytosis of latex beads (Fig. 4h). In response to LPS stimulation in vitro, switch of the CD11b expression occurred on AMs as well. Gpr84−/− AMs showed lower CD11b level under LPS challenge (Fig. 4i). In summary, Gpr84 played an important role in the regulation of macrophage functions, particularly AMs, including cytokine release, phagocytosis and status switch in LPS-induced inflammatory reactions.
In view of downregulated LPS-induced inflammatory responses of Gpr84−/− AMs, we assumed that GPR84 downstream may share crosstalk with TLR4-related pathways, instead of regulating single inflammatory cytokine. Lower expressions of Tlr4, MD-2, Cd14, and Lbp in LPS-induced Gpr84−/− AMs supported this assumption (Fig. 5a). CD14 was also downregulated in Gpr84−/− AMs under LPS challenge, compared with WT, whilst TLR4-MD2 dimers showed no significant difference (Fig. 5b). With intratracheal instillation of FITC-conjugated LPS in WT and Gpr84−/− mice, we explored that Gpr84−/− AMs exhibited weaker LPS-binding capacity in vivo (Fig. 5d). LPS-induced upregulated CD14 level, not TLR4/MD2 dimers, was reduced on Gpr84−/− AMs in vivo (Fig. 5c), which verified that CD14 level of AMs was regulated by Gpr84 in some way. Meanwhile, the ratio of LPS-binding CD14+ AMs in BALFs declined in Gpr84−/− mice (Fig. 5e). LBP release was also decreased in BALF from Gpr84−/− mice, implying that Gpr84 was involved in the regulation of LPS-mediated TLR4-related pathway via CD14 and LBP (Fig. 5e). Other pulmonary cells showed no significant difference in LPS-binding capacity between WT and Gpr84−/− mice (Supplementary Fig. 4b).
a Expressions of Tlr4, MD-2, Cd14 and Lbp in Gpr84+/+ and Gpr84−/− AMs with LPS challenge in vitro using real-time qPCR; b CD14 level in Gpr84+/+ and Gpr84−/− AMs after 3 h LPS challenge in vitro; c CD14 level in Gpr84+/+ and Gpr84−/− AMs after 3 h post-instillation of LPS-FITC in vivo. d LPS-binding capacity of Gpr84+/+ and Gpr84−/− AMs in vivo; e CD14+LPS+ AMs and LBP release in BALF from Gpr84+/+ and Gpr84–/− mice. Data of a–e are shown as means ± SEM (four biological repeats, three technical repeats), *0.01 < P < 0.05; **P < 0.01; ***P < 0.001 .
Gpr84 positively regulates CD14 and LBP via Akt/Erk1/2-STAT3
The regulatory mechanisms of GPR84 related to CD14 and LBP were next investigated. Due to limited number of isolated AMs, it was difficult to acquire sufficient AM proteins for western-blot assays. Instead, primary BMDMs were used to explore the GPR84 downstream pathways in macrophages. Gpr84−/− BMDMs showed lower phosphorylated levels of PI3K, Akt, Src, Erk1/2, p38 MAPK, STAT1, STAT3, and NF-κB (Fig. 6a, b) under LPS challenge within 1 h, the time point that expressions of Cd14 and Lbp were upregulated. Interestingly, β-arrestin 2, a negative regulator of GPCR signaling, was higher in Gpr84−/− BMDMs at basal level and maintained throughout LPS challenge period, whilst β-arrestin 1 showed no significant difference (Fig. 6b). This may be a key regulatory mechanism of Gpr84 deficiency in inflammation. Consistent with those found in BMDMs, obvious reduction in phosphorylated Akt, Erk1/2, and STAT3 were affirmed in Gpr84−/− AMs with Phosflow assays (Fig. 6d) and Confocal assays (Fig. 7a–c). Blockade of PI3K, Akt, STAT1, STAT3, ERK1/2, NF-κB, and p38 MAPK were performed with inhibitors to clarify their involvement in the regulation of CD14 and LBP (Fig. 7d). Treatment with Akt and STAT3 inhibitors downregulated LPS-induced CD14 and LBP of AMs in both mRNA and protein levels, whilst Erk inhibitors seemed to only suppress LBP level. The binding sites of STAT3 and promotor regions of Cd14 and Lbp were checked with ChIP assay (Fig. 6c). Combination of Phosflow, Confocal, inhibitor treatment, and ChIP suggested that Gpr84 regulated CD14 and LBP via Akt/Erk1/2-STAT3.
a WB assay for signal proteins in Gpr84+/+ and Gpr84−/− BMDMs under LPS challenge within 1 h; b WB assay for signal proteins in Gpr84+/+ and Gpr84−/− BMDMs under LPS challenge within 20 min; c ChIP analysis for binding of STAT3 to the promotor site of Cd14 and Lbp in BMDMs; d Phosflow analysis for phosphorylated level of signal proteins in Gpr84+/+ and Gpr84−/− AMs. Data of c are shown as means ± SEM (two biological repeats, three technical repeats), data of d are shown as means ± SEM (four biological repeats, three technical repeats), *0.01 < P < 0.05; **P < 0.01; ***P < 0.001.
a–c Confocal assays for phosphorylated levels of Akt, Erk1/2, and STAT3 in Gpr84+/+ and Gpr84−/− AMs under LPS challenge; d Effects of signal protein inhibitors on the CD14 expression and LBP release in primary AMs, including PI3K (LY294002), Akt (GSK2141795), STAT1 (Fludarabine), STAT3 (S3I201), NF-κB (JSH-23), Erk1/2 (GDC0994), and p38 MAPK (SB203580) (section symbol indicates (§) compared with PBS group, hash symbol indicates (#) compared with DMSO group, asterisk symbol indicates (*) compared with LPS + DMSO group). Data of d are shown as means ± SEM (four biological repeats, two technical repeats), *0.01 < P < 0.05; **P < 0.01; ***P < 0.001.
Discussion
Resident AMs and myeloid-derived MO/Mφ are both essential for the pathogenesis of ARDS/ALI, but the underlying regulatory mechanisms still need to be elucidated. Contradictory views on AMs in ARDS/ALI have been reported, which may imply different regulatory roles of AMs in different phases of pathogenesis. Here we would like to discuss about CD11bhi AMs, GPR84 and the potential immunometabolic mechanisms involved.
CD11bhi macrophages have been discovered in lung diseases long ago. With respect to the origin of CD11bhi macrophages, some regarded them as recruited monocytes,69 whilst others considered as a switched status from CD11blo AMs under challenge.37 In this study, we found that both CD11bhi AMs and recruited MO/Mφ existed in ALI. Recruited Ly-6G−CD45.2+F4/80+CD11a+Siglec-F−Ly-6Cmid-hiCD11clo-midCCR2mid-hiCX3CR1lo-midCD14hiCD326−CD11bhi MO/Mφ were discovered in both BALF and parenchyma cells, but Ly-6G−CD45.2+F4/80+CD11a+Siglec-F+Ly-6CmidCD11c+CCR2loCX3CR1lo-midCD14lo-midCD326lo-midCD11blo-hi AMs coupled with dynamic switch from CD11blo to CD11bhi status still dominated the F4/80+ macrophages in alveolar niches.
Several studies have shown that myeloid-derived MO/Mφ and some tissue-resident Mφ have the potential to participate in AM repopulation. With chimeric BMT, we confirmed that myeloid-derived MOs can repopulate the AMs under stress of lethal irradiation. With PMT, we demonstrated that M-CSF-induced mature BMDMs can also be transformed to an AM-like phenotype in alveolar niches in vivo and act as AMs. Only clodronate-induced AM depletion without transplantation showed self-repopulation in a week post depletion, while the repopulated macrophages showed comparable level of AM markers to resident AMs, in agreement with previous studies.40 Combined with these studies and our data, we thought myeloid-derived MO/Mφ, mature BMDMs, and resident AMs all have the repopulation potential, which may be related to the restricted alveolar niches and the status of resident AMs. The local proliferation of resident AMs may be more preferable, however, in pathological states with severe AM apoptosis or necrosis, such as lethal irradiation or drug-induced necrosis, MO/Mφ with different phenotypes can also contribute to the AM repopulation.
With regards to the characteristics of CD11bhi AMs, some hold the opinion that CD11bhi AMs are M2-like macrophages,70 whilst CD11b negatively regulates TLR4-mediated inflammation71 and has positive effects on DCs.72 Others believe that CD11b inhibition on AMs can alleviate inflammation.73 As far as we are concerned, CD11bhi AMs cannot be exclusively classified as a simple pro- or anti-inflammatory status, partly based on the consistently upregulated mRNA levels of macrophage activation markers, which was in agreement with recent studies.69 Elevated secretion of TNF, IL-6, IL-12 p40/p70, and enhanced phagocytic capacity of CD11bhi AMs revealed CD11bhi AMs as a more inflamed status, particularly in the inflammatory response phase of ARDS/ALI. Combination of increased CD11b expression on AMs under LPS challenge and the inflamed phenotype of CD11bhi AMs can simply imply CD11b as a hallmark of inflammatory status, regardless of CD11b functions. Our results did not mean to directly disprove the negative regulatory role of CD11b on AMs, since AMs may show the phenotypes that promote tissue repair in later phases. The underlying functions of the CD11bhi AMs will be analyzed furthermore.
Gpr84 was first discovered in neutrophils, the main invaded cells and source of reaction oxygen species production in ARDS/ALI. Interestingly, GPR84 effect on chemotaxis of neutrophils did exist but seemed not obvious as previously described, based on our results of chimeric BMT, PMT, and trans-well assay (Supplementary Fig. 3d–f). Chemotaxis ratio in several gated cells suggested that GPR84 may play opposed roles in chemotaxis of myeloid-derived cells and lymphocytes. Besides, CD45.1+WT neutrophils exhibited minor advantage of chemotaxis over CD45.2+Gpr84−/− neutrophils. PMT in vivo and trans-well assay in vitro also verified the results of chimeric BMT.
Different members of FFARs have been reported diverse important roles in diseases. However, the roles of GPR84 are still incompletely described. In our opinion, the ligands of GPR84 still remain uncharacterized. MCFAs (C9–C14, especially C10–C12), which were recognized as endogenous ligands of GPR84, can merely activate GPR84 at mM level concentrations in vitro. Since the release and forms of MCFAs in vivo remain a black box, we prefer an intermediate derivatives of MCFAs, such as derivatives with hydroxyl, which showed more effective to activate GPR84 in vitro.
Pulmonary immune and metabolic homeostasis were both disrupted in ARDS/ALI. We hypothesized that metabolic pathways of FFAs in the lung microbiota were disordered, leading to the accumulation of intermediate derivatives of MCFAs (C9–C14) in the microbiota and release into alveolar niches through exosomes or other vehicles. Recent studies have suggested that LPS stimulation can increase triglyceride accumulation and FFAs uptake of macrophages.52 However, the precise mechanisms by which FFAs and their derivatives generated from the microbiota contributing to ARDS/ALI or other lung diseases remain undefined, and require investigation in future studies.
In summary, our work reveals a key role as well as possible mechanisms of Gpr84 in regulating AMs in the ALI mouse model, including switch of CD11b status, acute inflammatory cytokine release, and phagocytic capacity. Our findings amplify the knowledge of different macrophage populations in pulmonary inflammatory responses. Involvement of FFARs in ARDS/ALI indicates an important role of immunometabolism in pulmonary pathogenesis. The regulatory mechanisms of pulmonary immune homeostasis, metabolic homeostasis, and their crosstalk need to be clarified depth.
Methods
Animals
Wild-type C57BL/6 mice were purchased from the Shanghai Laboratory Animal Company (Shanghai, China). Gpr84 deficient C57BL/6 mice and C57BL/6-Ly5.1 (CD45.1) mice were obtained from the Laboratory Animal Center of East China Normal University (Shanghai, China). Gpr84 deficient mice and CD45.1 mice were identified by PCR and FCM, respectively. All mice were bred and housed under specific pathogen-free (SPF) conditions according to institutional guidelines. Mice were sacrificed by cervical dislocation. The experimental protocols were approved by the Animal Investigation Committee of East China Normal University.
LPS-induced ALI mouse models and LPS interactions in vivo
Weight-matched male littermates (24–26 g) of different genotypes were used for LPS-induced ALI mouse models. Mice were anesthetized and administered with 1 mg/kg LPS (L2880, Sigma) by intratracheal instillation. Sham groups were given an identical volume of sterilized PBS. For the detection of LPS interactions in vivo, 4 mg/kg FITC-conjugated LPS (F8666, Sigma) was administered by intratracheal instillation.
AM depletion
PBS- or clodronate-liposomes (CP-005–005, LIPOSOMA) in a volume of 70 μL were administered by intratracheal instillation. The efficiency of depletion was measured after 24 h post injection. LPS-induced ALI models were established for a further 48 h.
Chimeric bone-marrow transplantation (BMT)
Age-matched male mice (8–10 weeks) of different genotypes were used. 1 g L−1 Ampicillin (A8180, Solarbio) and 1 g L−1 Neomycin (4801, Sigma) were added to the drinking water 1 week before BMT and 2 weeks post BMT. Recipient mice were irradiated at dose of 9 Gys. BM cells from CD45.1+ WT and CD45.2+ Gpr84−/− were isolated, counted, and mixed with different ratios (2:1 or 1:2) before injection. 5 × 106 chimeric BM cells in a total volume of 150 μL PBS were transplanted to each recipient mice via caudal vein. Mice were ready after 8 weeks post BMT.
For tracing of AM development, 5 × 106 chimeric BM cells with 1:1 chimeric ratio were transplanted. Mice were sacrificed from 1st to 8th week post transplantation. Both BALF cells and BM cells were isolated and analyzed.
Pulmonary macrophage transplantation (PMT)
PMT was performed as described previously.34 Age-matched male mice (8–10 weeks) of different genotypes were used. AMs of recipient mice were temporarily depleted with clodronate liposomes for consecutive 7 days. Donor BMDMs were obtained as described below. BMDMs were digested with 0.5% Trypsin-EDTA (15400054, Gibco) and counted. 2 × 106 BMDMs in a total volume of 50 μL PBS were administrated by intratracheal instillation. Mice were ready after 4 weeks post PMT.
Isolation of bronchoalveolar lavage fluid (BALF) cells and lung parenchyma cells
Mice were euthanatized. BALF cells were collected by intratracheal instillation of pre-warmed PBS and temporarily stored on ice. Lung tissues were then sectioned and digested with 1 mg mL−1 Collagenase D (11088858, Roche), 1 mg mL−1 Dispase II (04942078001, Roche), and 100 U mL−1 DNase I (D5025, Sigma) at 37 °C for 1 h. Digested samples were passed through 40 μm cell meshes and temporarily stored on ice. All samples were centrifuged and BALF supernatants were collected for ELISA. RBCs were lysed and residual cell pellets were resuspended with precooled MACS solution (D-PBS containing 2% heat-inactivated FBS and 0.25 mM EDTA) for FCM analysis.
FCM analysis and cell sorting
For surface markers, cells were blocked for CD16/32 with functional-grade antibody (16–0161, eBioscience) for 15 min at 4 °C. Surface markers were stained for another 1 h at 4 °C, washed and resuspended with MACS solution.
For intracellular staining, cells were isolated in sterile conditions and plated in 24-well multidishes. Non-adherent cells were discarded 2 h later. Adherent cells were primed with 20 ng mL−1 IFN-γ (315–05, PEPROTECH) for 2 h and stimulated with 1 μg mL−1 LPS and 2 μL protein transport inhibitor cocktail (eBioscience, 00–4980) for another 12 h. Cells were digested with 0.5% Trypsin-EDTA, centrifuged and resuspended. Following CD16/32 blockade and staining of specific markers, cells were fixed and permeabilized using BD Fixation and Permeabilization Kits (554714, BD).
For phosflow, AMs were isolated and incubated at 37 °C overnight. Phosflow were performed as BD protocol described. Single cell suspensions of AMs were prepared the second day and recovered in water bath at 37 °C for 2 h. Cells were treated with PBS or LPS, fixed with BD Phosflow Lyse/Fix Buffer (558049, BD), and permeabilized with BD Phosflow Perm/Wash Buffer I (557885, BD).
All analysis (Table 1) were performed with a BD LSRFortessa and FlowJo X.
For cell sorting, BALF and lung parenchyma cells were isolated in sterile conditions and stained for membrane markers. Cells were sorted with a BD Aria II and collected in complete medium.
Histology and immunohistochemistry (IHC)
Murine lungs were fixed with 4% paraffin overnight. Samples were dehydrated with dose-gradient ethanol, immersed in xylene for 20 min, and embedded with paraffin. Slices (5 μm) were prepared and stored at −20 °C for further HE staining and IHC at Servicebio (Shanghai, China).
Lung permeability assay
20 mg/kg Evans blue stain (E2129, Sigma) was injected through the caudal vein of mice. Blood samples were collected from the posterior orbital plexus with capillary after 30 min post injection. Blood samples were clarified at 16,000 g for 5 min at 4 °C. Supernatants were removed and stored at −80 °C. Mice were euthanized by cervical dislocation. Lung tissue samples were dissociated, washed with PBS, and freeze-dried at −80 °C. Frozen samples were weighed, homogenized with 500 μL formamide, and incubated at 56 °C for 48 h. Mixtures were clarified at 5,000 g for 30 min and transferred to new tubes. Evans blue concentrations were assessed based on the optical density of the samples at 620 and 740 nm. For blood samples, data were corrected with the formula: E620 (corrected) = E620 − (1.426 × E740 + 0.030). For lung tissue samples, values were measured with E620 and normalized by tissue weight.
ELISA
TNF, IL-6, and CCL2 were measured with ELISA Max Standard Sets (430904, 431301, 432701, BioLegend), IL-12 were measured with ELISA Ready-SET-GO (88–7121, eBioscience). All ELISA measurements followed standard steps recommended on TDS kits.
RNA extraction and reverse transcription PCR
For isolated cells and tissues, RNA was extracted with RNAiso Plus (9108, TaKaRa) and reverse transcribed with PrimeScript™ RT reagent kits (RR036A, TaKaRa). For sorted cells, RNA was extracted and reverse transcribed with Cell-to-cDNA Kits (B0001, EZ Bioscience).
RNA interference (RNAi)
MH-S cells were plated in the absence of Penicillin or Streptomycin. Media was replaced with OptiMEM when the cell density was about 60–70%. Cells were then treated with a mixture of siGpr84 (sense: 5′-UCUGUGUUGGGCUAUCGAUTT-3′, antisense: 5′-AUCGAUAGCCCAACACAGATT-3′, GenePharma, Shanghai) and Lipofectamine RNAiMAX Reagent (13778–100, Invitrogen) for 6 h. Cells were stimulated after 24 h post transfection.
Isolation of AMs BMDMs, PEMs in vitro
Mice were euthanatized by cervical dislocation and dipped in 75% EtOH. To isolate AMs and BMDMs, lung tissues and hind legs were separated and gently washed in sterilized PBS. For AMs, pre-warmed Ca2+, Mg2+-free D-PBS containing 0.5 mM EDTA was infused into alveolar niches and collected. Suspensions were mixed with 1 mL of complete RPMI 1640 medium to neutralize the EDTA. For BMDMs, marrow cavities were flushed with serum-free DMEM until visible. Suspensions were then passed through 40 μm cell meshes.
Cell suspensions from alveolar niches and bone cavities were centrifuged at 300 g for 3 min and supernatant was discarded. RBC lysis were performed in BD Lyse (555899, BD) for 5 min at room temperature and centrifuged. Supernatants were discarded and cell pellets were resuspended in complete RMPI 1640 (for AMs) or DMEM (for BMDMs) containing 10% heat-inactivated FBS and 1% P/S (15140122, Gibco). For AMs, the media were replenished after 2 h and cells were cultured overnight for further analysis. For BMDMs, non- and weakly-adherent cells were added to six-well plates the second day and cultured in complete DMEM and 20 ng mL−1 M-CSF (315–02, PEPROTECH) for 7 days, with exchange of media at 3rd and 6th day.
To isolate PEMs, 3 mL thioglycollate medium (70157, Millipore) was injected i.p. for successive 3 days in SPF conditions. The peritoneum was exposed and filled with 2 mL D-PBS containing 0.5 mM EDTA. Peritoneal fluid was withdrawn and temporarily stored on ice. Cell suspensions were centrifuged and RBCs were lysed. Media were replaced 2 h later and adherent cells were incubated with complete DMEM overnight and ready for further analysis.
Analysis of phagocytic capacity
AMs were pretreated with PBS or LPS. FITC-conjugated latex beads (L1030, Sigma) were 1:1000 diluted with PBS and added to the media. Supernatant was discarded 1 h later. Cells were washed, digested and analyzed with BD LSRFortessa and FlowJo X.
Isolation of neutrophils and trans-well assay
BM cells were isolated as described above. Separation column with gradient density of 45, 62, 81% was made with Percoll (17–0891–01, GE Healthcare) and Ca2+, Mg2+-free HBSS (14175–095, Gibco). BM cells were resuspended with 45% media, added to the column and centrifuged at 1,500 g for 30 min at 4 °C with low accelerating and descending rate. Cells were gently obtained, resuspended with serum-free RPMI 1640, and centrifuged at 300 g for 5 min at 4 °C. Cell pellets were resuspended with complete RPMI 1640 with 25 ng mL−1 GM-CSF (567302, BioLgend) overnight and ready for further analysis.
For trans-well assay, AMs were cultured in lower chambers overnight. Media were exchanged with complete RPMI 1640 containing 2% heat-inactivated FBS and 1% P/S just prior to the experiment. AMs were stimulated with 1 μg mL−1 LPS for 6 h. f-MLP (1921, TOCRIS) were used as positive chemoattractant. Neutrophils were collected, centrifuged, and resuspended with complete RPMI 1640 containing 2% heat-inactivated FBS and 1% P/S at a density of 3 × 105 cells per 200 μL. Resuspended cells in a volume of 200 μL were added to upper chambers (353504, BD Falcon) and migrated for 2 h. Cell suspensions in lower chambers were collected, stained, and analyzed for Ly-6G+ cells in 1 min with BD LSRFortessa and FlowJo X.
Real-time quantitative PCR
TB Green™ Premix Ex Taq™ (RR420A, TaKaRa) and SimpleChIP Universal qPCR Master Mix (88989, CST) were used. Real-time quantitative PCR (Tables 2, 3) was performed on the QuantStudio™ 3 Real-Time PCR System (A28566, Applied Biosystems).
Western blot (WB)
Cells were lysed with RIPA solution containing 1% protease inhibitor cocktail (CW2200S, CWBIO) and 1% Phosphatase inhibitor cocktail (CW2383, CWBIO). Lysed mixture were sonicated and quantified with BCA Protein Assay Kit (A53225, Thermo Scientific). Normalized samples were boiled with SDS-PAGE buffer (7722, CST) and stored at −80 °C. SDS-PAGE was performed at 80 V and gel was transferred at 300 mA. Membranes were blocked with milk, washed with PBST, and incubated with primary antibody (Table 4) overnight at 4 °C. Membranes were washed, incubated with secondary antibody for 1 h at room temperature, washed, and scanned with Odessey CLX.
Immunofluorescence (IF) and confocal assay
AMs were isolated as described above and cultured on coverslips. After treatment with PBS or LPS, media were discarded. AMs were fixed with 4% paraffin for 15 min at room temperature, washed with TBST, and permeabilized successively with methanol, 1% BSA, and 0.3% Triton X-100. Supernatant was then discarded and cells were incubated with primary antibodies (Table 4) overnight at 4 °C. Samples were washed, incubated with secondary antibody or phalloidin-iFluor 555 (ab176756, Abcam) at room temperature for 40 min and washed again. Coverslips were mounted on slides with SlowFade Diamond Antifade Mountant (S36968, Invitrogen), scanned with Lecia TCS SP8, analyzed with Lecia Application Suite X and merged with ImageJ.
Chromatin Immunoprecipitation (ChIP)
ChIP were performed with SimpleChIP Plus Enzymatic Chromatin IP Kit (9005, CST). Cells were cross-linked with 37% formaldehyde (F8775, Sigma) and performed as CST protocol described. Samples from BMDMs were treated with 0.25 μL Micrococcal Nuclease for 5 min at 37 °C. Transcription factor binding sites (TFBS) were predicted with JASPAR (jaspar.genereg.net) and hTFtarget (bioinfo.life.hust.edu.cn/hTFtarget). Primers were designed with Primer Premier 6 and evaluated for the primer specificity with Primer-BLAST.
Statistical analysis
Data are presented as the mean ± SEM. All statistical analysis were performed with GraphPad Prism version 7.0. Significant differences were assessed through t test for comparisons between groups and a one-way ANOVA for comparisons amongst more than two groups.
References
Sweeney, R. M. & McAuley, D. F. Acute respiratory distress syndrome. Lancet 388, 2416–2430 (2016).
Thompson, B. T. et al. Acute respiratory distress syndrome. N. Engl. J. Med. 377, 562–572 (2017).
Arndt, P. G., Fantuzzi, G. & Abraham, E. Expression of interleukin-18 in the Lung after Endotoxemia or Hemorrhage-Induced acute lung injury. Am. J. Respir. Cell Mol. Biol. 22, 708–713 (2000).
Gibbs, J. et al. An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat. Med. 20, 919–926 (2014).
Murata, T. et al. Anti-inflammatory role of PGD2 in acute lung inflammation and therapeutic application of its signal enhancement. Proc. Natl Acad. Sci. USA 110, 5205–5210 (2013).
Matthay, M. A. & Zemans, R. L. The acute respiratory distress syndrome: pathogenesis and treatment. Annu. Rev. Pathol. 6, 147–163 (2011).
Herold, S. et al. Acute lung injury: how macrophages orchestrate resolution of inflammation and tissue repair. Front. Immunol. 2, 65 (2011).
Peters, D. M. et al. TGF-beta directs trafficking of the epithelial sodium channel ENaC which has implications for ion and fluid transport in acute lung injury. Proc. Natl Acad. Sci. USA 111, E374–E383 (2014).
Zacharias, W. J. et al. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251–255 (2018).
Hussell, T. & Bell, T. J. Alveolar macrophages: plasticity in a tissue-specific context. Nat. Rev. Immunol. 14, 81–93 (2014).
Perdiguero, E. G. & Geissmann, F. The development and maintenance of resident macrophages. Nat. Immunol. 17, 2–8 (2016).
Westphalen, K. et al. Sessile alveolar macrophages communicate with alveolar epithelium to modulate immunity. Nature 506, 503–506 (2014).
Okabe, Y. & Medzhitov, R. Tissue biology perspective on macrophages. Nat. Immunol. 17, 9–17 (2016).
Hartl, D. et al. Infiltrated neutrophils acquire novel chemokine receptor expression and chemokine responsiveness in chronic inflammatory lung diseases. J. Immunol. 181, 8053–8067 (2008).
Nandi, B. & Behar, S. M. Regulation of neutrophils by interferon-gamma limits lung inflammation during tuberculosis infection. J. Exp. Med. 208, 2251–2262 (2011).
Kolaczkowska, E. & Kubes, P. Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 13, 159–175 (2013).
Schmidt, E. P. et al. The pulmonary endothelial glycocalyx regulates neutrophil adhesion and lung injury during experimental sepsis. Nat. Med. 18, 1217–1223 (2012).
Laidlaw, B. J. et al. CD4+ T cell help guides formation of CD103+ lung-resident memory CD8+ T cells during influenza viral infection. Immunity 41, 633–645 (2014).
Thawer, S. G. et al. Lung-resident CD4(+) T cells are sufficient for IL-4Ralpha-dependent recall immunity to Nippostrongylus brasiliensis infection. Mucosal Immunol. 7, 239–248 (2014).
Mock, J. R. et al. Foxp3+ regulatory T cells promote lung epithelial proliferation. Mucosal Immunol. 7, 1440–1451 (2014).
John-Schuster, G. et al. Cigarette smoke-induced iBALT mediates macrophage activation in a B cell-dependent manner in COPD. Am. J. Physiol. Lung Cell. Mol. Physiol. 307, L692–L706 (2014).
Martinez-Gonzalez, I. et al. ILC2 memory: recollection of previous activation. Immunol. Rev. 283, 41–53 (2018).
Whitsett, J. A. & Alenghat, T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat. Immunol. 16, 27–35 (2015).
Holtzman, M. J. et al. The role of airway epithelial cells and innate immune cells in chronic respiratory disease. Nat. Rev. Immunol. 14, 686–698 (2014).
Weitnauer, M. et al. Control of local immunity by airway epithelial cells. Mucosal Immunol. 9, 287–298 (2016).
Nikota, J. K. et al. Cigarette smoke primes the pulmonary environment to IL-1alpha/CXCR-2-dependent nontypeable Haemophilus influenzae-exacerbated neutrophilia in mice. J. Immunol. 193, 3134–3145 (2014).
Steele, C. et al. The beta-glucan receptor dectin-1 recognizes specific morphologies of Aspergillus fumigatus. PLoS Pathog. 1, e42 (2005).
Fehrenbach, H. et al. Alveolar macrophages are the main source for tumour necrosis factor-alpha in patients with sarcoidosis. Eur. Respir. J. 21, 421–428 (2003).
Farley, K. S. et al. Effects of macrophage inducible nitric oxide synthase in murine septic lung injury. Am. J. Physiol. Lung Cell Mol. Physiol. 290, L1164–L1172 (2006).
Goritzka, M. et al. Alveolar macrophage-derived type I interferons orchestrate innate immunity to RSV through recruitment of antiviral monocytes. J. Exp. Med. 212, 699–714 (2015).
Dagvadorj, J. et al. Lipopolysaccharide induces alveolar macrophage necrosis via CD14 and the P2X7 receptor leading to interleukin-1alpha release. Immunity 42, 640–653 (2015).
Coleman, M. M. et al. Alveolar macrophages contribute to respiratory tolerance by inducing FoxP3 expression in naive T cells. Am. J. Respir. Cell Mol. Biol. 48, 773–780 (2013).
Soroosh, P. et al. Lung-resident tissue macrophages generate Foxp3+ regulatory T cells and promote airway tolerance. J. Exp. Med. 210, 775–788 (2013).
Suzuki, T. et al. Pulmonary macrophage transplantation therapy. Nature 514, 450–454 (2014).
Dong, Y. et al. The survival of fetal and bone marrow monocyte-derived alveolar macrophages is promoted by CD44 and its interaction with hyaluronan. Mucosal Immunol. 11, 601–614 (2018).
Nakamura, A. et al. Transcription repressor Bach2 is required for pulmonary surfactant homeostasis and alveolar macrophage function. J. Exp. Med. 210, 2191–2204 (2013).
Huaux, F. et al. IL-1alpha induces CD11b(low) alveolar macrophage proliferation and maturation during granuloma formation. J. Pathol. 235, 698–709 (2015).
Guilliams, M. et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J. Exp. Med. 210, 1977–1992 (2013).
Lavin, Y. et al. Regulation of macrophage development and function in peripheral tissues. Nat. Rev. Immunol. 15, 731–744 (2015).
Hashimoto, D. et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38, 792–804 (2013).
Sieweke, M. H. & Allen, J. E. Beyond stem cells: self-renewal of differentiated macrophages. Science. 342, 1242974 (2013).
Van den Bossche, J. et al. Macrophage immunometabolism: where are we (Going)? Trends Immunol. 38, 395–406 (2017).
O’Neill, L. A. & Pearce, E. J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 213, 15–23 (2016).
Gaber, T. et al. Metabolic regulation of inflammation. Nat. Rev. Rheumatol. 13, 267–279 (2017).
Ganeshan, K. & Chawla, A. Metabolic regulation of immune responses. Annu. Rev. Immunol. 32, 609–634 (2014).
Divakaruni, A. S. et al. Etomoxir inhibits macrophage polarization by disrupting CoA homeostasis. Cell Metab. 28, 490–503.e7 (2018).
Huang, L. et al. Growth of mycobacterium tuberculosis in vivo segregates with host macrophage metabolism and ontogeny. J. Exp. Med. 215, 1135–1152 (2018).
Xie, N. et al. Metabolic characterization and RNA profiling reveal glycolytic dependence of profibrotic phenotype of alveolar macrophages in lung fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 313, L834–l44 (2017).
Potter, B. J. et al. Mechanisms of cellular uptake of free fatty acids. Annu. Rev. Nutr. 9, 253–270 (1989).
Huang, S. C. et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat. Immunol. 15, 846–855 (2014).
Man, W. H. et al. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat. Rev. Microbiol. 15, 259–270 (2017).
Feingold, K. R. et al. Mechanisms of triglyceride accumulation in activated macrophages. J. Leukoc. Biol. 92, 829–839 (2012).
Berod, L. et al. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat. Med. 20, 1327–1333 (2014).
Yao, C. H. et al. Exogenous fatty acids are the preferred source of membrane lipids in proliferating fibroblasts. Cell Chem. Biol. 23, 483–493 (2016).
Pan, Y. et al. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature 543, 252–256 (2017).
Tan, J. K. et al. Metabolite-sensing G protein-coupled receptors-facilitators of diet-related immune regulation. Annu. Rev. Immunol. 35, 371–402 (2017).
Thorburn, A. N. et al. Diet, metabolites, and “western-lifestyle” inflammatory diseases. Immunity 40, 833–842 (2014).
Bloes, D. A. et al. Enemy attraction: bacterial agonists for leukocyte chemotaxis receptors. Nat. Rev. Microbiol. 13, 95–104 (2015).
Fujita, T. et al. A GPR40 agonist GW9508 suppresses CCL5, CCL17, and CXCL10 induction in keratinocytes and attenuates cutaneous immune inflammation. J. Investig. Dermatol. 131, 1660–1667 (2011).
Oh, D. Y. et al. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 142, 687–698 (2010).
Yousefi, S. et al. Cloning and expression analysis of a novel G-protein-coupled receptor selectively expressed on granulocytes. J. Leukoc. Biol. 69, 1045–1052 (2001).
Wang, J. et al. Medium-chain fatty acids as ligands for orphan G protein-coupled receptor GPR84. J. Biol. Chem. 281, 34457–34464 (2006).
Bouchard, C. et al. G protein-coupled receptor 84, a microglia-associated protein expressed in neuroinflammatory conditions. Glia 55, 790–800 (2007).
Nagasaki, H. et al. Inflammatory changes in adipose tissue enhance expression of GPR84, a medium-chain fatty acid receptor: TNFalpha enhances GPR84 expression in adipocytes. FEBS Lett. 586, 368–372 (2012).
Suzuki, M. et al. Medium-chain fatty acid-sensing receptor, GPR84, is a proinflammatory receptor. J. Biol. Chem. 288, 10684–10691 (2013).
Nicol, L. S. et al. The role of G-protein receptor 84 in experimental neuropathic pain. J. Neurosci. 35, 8959–8969 (2015).
Recio, C. et al. Activation of the immune-metabolic receptor GPR84 enhances inflammation and phagocytosis in macrophages. Front. Immunol. 9, 1419 (2018).
Guilliams, M. & Scott, C. L. Does niche competition determine the origin of tissue-resident macrophages? Nat. Rev. Immunol. 17, 451–460 (2017).
Duan, M. et al. Distinct macrophage subpopulations characterize acute infection and chronic inflammatory lung disease. J. Immunol. 189, 946–955 (2012).
Wang, J. et al. Bacterial colonization dampens influenza-mediated acute lung injury via induction of M2 alveolar macrophages. Nat. Commun. 4, 2106 (2013).
Han, C. et al. Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating Syk and promoting degradation of MyD88 and TRIF via Cbl-b. Nat. Immunol. 11, 734–742 (2010).
Ling, G. S. et al. Integrin CD11b positively regulates TLR4-induced signalling pathways in dendritic cells but not in macrophages. Nat. Commun. 5, 3039 (2014).
Duan, M. et al. CD11b immunophenotyping identifies inflammatory profiles in the mouse and human lungs. Mucosal Immunol. 9, 550–563 (2016).
Acknowledgements
This work was supported by National Key R&D Program of China [2018YFA0507001]; National Nature Science Foundation of China [81871250, 81672811, 81771306]; Innovation Program of Shanghai Municipal Education Commission [2017-01-07-00-05-E00011]; Shanghai Sailing Program [19YF1414400]. We thank East China Normal University Multifunctional Platform for Innovation [011] for helping carrying out animal experiments.
Author information
Authors and Affiliations
Contributions
B.D., M.-y.L., Y.Z., and W.-z.J. provided advice and technical expertise. C.-c.Y., L.C., L.-l.C., and S.-y.W. performed experiments and analyzed data. J.-j.P., Q.X., and J.-l.Q. supervised the animal experiments performed in Scientific Laboratory Animal Center in East China Normal University. C.-c.Y. and H.R. wrote the paper. All authors revised the paper and approved its final version. M.Q. and H.R. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Yin, C., Cheng, L., Pan, J. et al. Regulatory role of Gpr84 in the switch of alveolar macrophages from CD11blo to CD11bhi status during lung injury process. Mucosal Immunol 13, 892–907 (2020). https://doi.org/10.1038/s41385-020-0321-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-020-0321-7
This article is cited by
-
Targeting metabolic sensing switch GPR84 on macrophages for cancer immunotherapy
Cancer Immunology, Immunotherapy (2024)
-
Regulator of G protein signaling protein 6 alleviates acute lung injury by inhibiting inflammation and promoting cell self-renewal in mice
Cellular & Molecular Biology Letters (2023)
-
Pro-phagocytic function and structural basis of GPR84 signaling
Nature Communications (2023)
-
Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84
Nature Communications (2023)
-
GPR84 signaling promotes intestinal mucosal inflammation via enhancing NLRP3 inflammasome activation in macrophages
Acta Pharmacologica Sinica (2022)