Abstract
Cowpea (Vigna unguiculata (L.)) is an important crop for food security in Senegal; therefore, understanding the genetic diversity of local germplasm is relevant for crop improvement and genetic maintenance in the era of climate change. For this purpose, 15 microsatellite markers were used to estimate the genetic diversity of Senegalese cowpea germplasm, including 671 accessions grown in eight regions and 66 wild relatives and intermediate forms (weedy). For the cultivated, the main expected heterozygosity (mHe) ranged between 0.317 (Fatick) and 0.439 (South). A narrow genetic variation between accessions from the different regions was observed with genetic similarity ranging from 0.861 to 0.965 and genetic differentiation indices (Fst) between 0.018 and 0.100. The accessions from southern Senegal (Kédougou, Sédhiou, and Kolda regions) are more diverse than the others. However, the accessions from the North (Saint-Louis) are genetically different from other regions. The diversity analysis in wild relatives from Senegal, which had never been performed before, revealed that the wild/weedy forms remain more diverse than the cultivated with genetic diversity values (He) of 0.389 and 0.480, respectively. STRUCTURE software divided the Senegalese germplasm into five subpopulations. Three of them (i, ii, and iii) included only cultivated accessions from several regions, one (v) mainly from Saint-Louis, and one (iv) the wild/weedy with some cultivated accessions. Our results support the hypothesis that Vigna unguiculata var. spontanea is the wild progenitor of cowpea. The accessions from the South, the northern recession accessions, and the wild/weedy could serve as sources of new genes for the genetic improvement of cowpea in Senegal.
Similar content being viewed by others
Introduction
Cowpea [Vigna unguiculata (L.) Walpers], a diploid crop (2n = 2x = 22), is one of the most important seed legumes in the arid and semi-arid zones of Africa with considerable economic, nutritional, and agronomic benefits (Agbicodo et al. 2009). The seeds are a valuable source of vegetable protein (25%) (Boukar et al. 2011) and are rich in lysine and tryptophan and serve as off-season food for rural populations. They also contain a significant amount of minerals and vitamins (folic acid and vitamin B), necessary for the prevention of congenital malformations (Hall et al. 2003; Diouf 2011). In addition, stalks and leaves of cowpea are used as forage in the Sahelian and Great Lakes regions (Pasquet and Baudoin 1997; Singh 2002; Cisse and Hall 2003). When dry, its biomass is used for animal feeding during the dry season when fresh grass becomes scarce in the Sudano-Sahelian zone. In addition to its nutritional qualities, cowpea has an economic benefit, providing significant supplementary income for rural populations when marketed. Agronomically, cowpea plays an important role as a source of nitrogen through its symbiotic association with Bradyrhizobium bacteria, reducing the demand of nitrogen fertilizer and the costs of agricultural production and is relatively drought-tolerant when compared to the other crops (Hall 2004). The world annual production of the crop is estimated at 5.7 million tons of dry seed (94% in Africa) with a relatively low average productivity of 505 kg/ha. In Africa, cowpea yields ranges between 448 and 578 kg/ha (FAOSTAT 2014). These weaknesses are due to several abiotic (heat, drought, salinity, etc.) and biotic (diseases, insects and parasitic plants) constraints (Obilana 1987; Singh and Sharma 1996). In Senegal, those constraints contribute to making cowpea yields even lower, ranging from 464 to 535 kg/ha with a production of 108,662 tons a year over on an estimated area of 63,857 ha (ANSD 2019).
To overcome these constraints, identifying sources of genetic diversity within local germplasm is an essential starting step towards genetic improvement, with the aim of limiting crop vulnerability to abiotic and biotic stresses and promoting conservation of important genetic resources (Barrett and Kidwell 1998).
In Senegal, breeding programs have been initiated since 1960 with the establishment of the first collection of cultivated cowpeas (Séne 1966). Another collection was also established by Kouakou et al. (2007), and genetic and morphological diversity studies have been conducted. However, these collections included cultivated varieties only from 3 Senegalese regions (Louga, Diourbel, and Thiès), and some of them have been lost over the time. Moreover, no collection of wild relatives has ever been carried out in Senegal despite the fact that these important species are exposed to genetic erosion. However, the sustainability of these programs is threatened by the narrow genetic basis of this legume. Various programs have suggested that the genetic basis can be broadened through the use of “omics” data or interspecific hybridization (Badiane et al. 2014). The latter method should be considered from very broad varietal collections, including cultivated accessions, landraces, and wild relatives which are the progenitors of cowpea. These genetic resources are important for boosting the effectiveness of cowpea improvement programs but are nowadays neglected in Senegal.
The diversity of cultivated cowpea was first assessed on the basis of morphological markers (Ehlers and Hall 1997). However, the low availability of morphological markers, the lack of knowledge on genetic control, and the influence of environmental factors on phenotypic expression at different stages of development are major limitations for using these markers as tools in diversity studies (Dikshit et al. 2007). In the past decades, biochemical markers, such as allozymes (Panella and Gepts 1992; Pasquet 1993) and seed storage proteins (Fotso et al. 1994) have been used to examine the genetic diversity of cultivated and wild cowpea. The biochemical markers have been abandoned, due to their limited number, sensitivity to environmental factors, and the developmental stage, in favor of markers based on DNA. Thus, chloroplast DNA polymorphisms (Vaillancourt and Weeden 1992), RAPD (Nkongolo 2003; Diouf and Hilu 2005), RFLP (Fatokun et al. 1993), DAF (Spencer et al. 2000; Simon et al. 2007), AFLP (Coulibaly et al. 2002), ISSR (Ghalmi et al. 2010), and SSR (Li et al. 2001; Ogunkanmi et al. 2008; Uma et al. 2009; Xu et al. 2010; Gupta and Gopalakrishna 2010; Asare et al. 2010; Lal et al. 2016; Desalegne et al. 2016) have been used to analyze the genetic variation among cowpea varieties. More recently, with the next generation sequencing (NGS), single nucleotide polymorphism (SNP) markers have been increasingly used in these types of studies and have been shown to be effective (Egbadzor et al. 2014; Desalegne et al. 2017). Nevertheless, SSRs remain extremely effective tools in diversity studies because, in addition to being abundant and randomly distributed in the genome, they are highly polymorphic, heritable, and co-dominant, easily reproducible and traceable with simple screening (Li Wang et al. 2008). Their use to analyze the genetic variation of some local and inbred varieties from Senegal show a low genetic diversity and have been limited to a few samples (Diouf and Hilu 2005; Kouakou et al. 2007; Badiane et al. 2012). Moreover, SSR markers allowed the evaluation of the genetic diversity of cowpea germplasm from Ghana (Asare et al. 2010) and Ethiopia and were also described as a powerful tool for identifying new genotypes within available collections and their use in crop selection and improvement programs (O’Neill et al. 2003).
To date, efforts have been made only to examine the genetic diversity of cultivated (landraces and improved) varieties in Senegal. Therefore, this study including the wild relative accessions was undertaken to address the lack of knowledge about Senegalese cowpea resources to improve the effectiveness of breeding programs and guide future germplasm conservation efforts.
Material and Methods
Plant Material
Cultivated cowpea were collected between September 2015 and March 2016 in eight regions in Senegal (Louga, Diourbel, Fatick, Thiès, Sédhiou, Kédougou, Saint-Louis, and Kolda). In each region, four to six villages were visited; for each village, nine to 11 randomly selected farmers were interviewed, and 671 accessions were collected (Table S1). Seed provenance, local name, sowing period, harvesting period, etc. were recorded during farmer interviews.
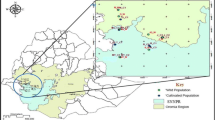
A second phase of prospecting and collecting the cowpea wild relative was carried out between September and December 2016. These missions allowed the establishment of a collection of 66 samples including 45 Vigna unguiculata var. spontanea and 21 intermediate forms (20 from the Midwest: Thiès, Louga, Diourbel, and Fatick and 1 from the South: Kolda) (Table S2). The geographical position of each collection site was recorded using Sygic GPS Navigation software (version 16) installed on a Samsung Tab 3V tablet, and the collection sites were mapped (Fig. 1).
In total, 737 samples including 671 cultivated cowpea and 66 wild relative and intermediate accessions from Senegal were used.
DNA Isolation and Genotyping
Genomic DNA was extracted from fresh leaves of 21-day-old seedlings from one plant per accession using the MATAB (mixed alkyl trimethyl ammonium bromide) method (Risterucci et al. 2000). The extracted DNA was quantified on a 1% agarose gel by concentration estimation and compared to the bands of a Smart Ladder (MW-1700-10-Eurogentec) of known concentration. The DNA of each accession was diluted to a concentration of 5 ng/μl.
Polymorphic microsatellite markers (Table S3) were selected after screening a batch of more than 300 cowpea markers available at Centre d’Etude Régional pour l’Amélioration de l’Adaptation à la Sécheresse (CERAAS). Among these markers, 15 polymorphic SSRs were selected and used for polymerase chain reaction (PCR) amplification (Table S3).
The PCR was performed using a 96-block thermal cycler (MWG AG biotech) in a total volume of 10 μl containing 25 ng of DNA, 1.1 μl PCR buffer (10×), 0.275 μl of Taq DNA polymerase (1 U), 0.5 mM MgCl2, 200 μM dNTPs, 0.09 μl of each forward and reverse primers, and M13 tail. PCR cycle conditions were as follows: 4 min initial denaturation at 94 °C, 35 cycles of amplification including 1 min at 94 °C, 1 min at 55 °C (for all primers except SSR6807 which was amplified at 50 °C), 1 min 15 s at 72 °C, and a final extension at 72 °C for 7 min.
PCR products were separated by electrophoresis on 6.5% acrylamide denaturing gel on Licor 4300 sequencer (LICOR Inc., NE, USA). The fragments marked during amplification emit fluorescence when excited by laser diodes at two different wavelengths (682 and 782 nm). An infrared camera detects signals, and images are automatically recorded and downloaded for analysis. Allele sizes were estimated by comparing with different bands of the size marker (ladder produced by CIRAD).
Diversity Analysis and Genetic Parameter Estimation
Data were checked with XnView 3.13 and Jelly 0.1 software. The alleles are coded in increasing order using the numbers from 1 to 15. The allele with the smallest size for each individual was assigned the number 1. Missing data have been materialized by 0.
For the first part, all accessions from a region represented a population, except those from the South, which is the area with the lowest cowpea production. In Sédhiou, Kolda, and Kédougou regions, a small number of samples of 73, 6, and 15, respectively, were collected. Therefore, all accessions from these three regions were considered as a population.
To assess the structure of genetic diversity, basic statistics were estimated. Numbers of alleles, genetic diversity (expected heterozygosity) (Nei 1978), fixation indices, genetic similarities, and molecular variance between accessions of regions were calculated for each SSR locus and populations (regions) using GenAlex 6.503 software (Peakall and Smouse 2012). The differentiation indexes (Fst) and the probability values between accessions of populations were calculated using adegenet package for R software (Jombart 2008).
Population structure was estimated using DARwin (Perrier and Jacquemoud-Collet 2016) and STRUCTURE software (Pritchard et al. 2000). STRUCTURE analysis was performed with K ancestral populations ranging from 1 to 10. We used 200,000 iterations and a burn-in period of 20,000; 10 runs for each K value were performed. Outputs were summarized using Structure Harvester (Earl and vonHoldt 2012). We evaluated the number of population K based on Evanno’s method (Evanno et al. 2005). For ancestry analysis (q), we used the simulation with the highest log probability. We classified individuals in groups based on an ancestry coefficient of 0.55 or higher.
Principal coordinate analysis (PCoA) was performed to represent the spatial distribution of individuals from different populations using the adegenet package for R software (Jombart 2008).
Results
Genetic Diversity of Senegalese Cowpea
Genetic Diversity of Cultivated Cowpea
The 15 SSR markers used to evaluate the genetic diversity in this study were polymorphic for detecting a total of 93 alleles. The detected number of alleles ranged from two (2) alleles (SSR6243, SSR6311, and SSR6217) to 15 (SSR6800 and SSR6807) with an average of 6.2 alleles per marker (Table 1).
In this study, the used SSR markers revealed high percentages of polymorphic loci ranging from 73% (Fatick) to 100% (Thiès region and southern Senegal) with an average of 91.11% (Table 3). The number of alleles for the six populations ranged from 52 (in Fatick region) to 77 (in the South). The mean expected heterozygosity (mHe) was low and ranged from 0.317 (in Fatick region) to 0.439 (in the South) with an average of 0.359. The most important diversity of cowpea was observed in the southern part of Senegal followed by Thiès region (mHe = 0.385), while the lowest diversity was noticed in Fatick (Table 2).
Genetic similarity values were high and ranged from 0.861 (between Saint-Louis and Thiès accessions) to 0.965 (between Thiès and southern accessions). Important similarities were observed between the accessions collected in the South region with those of the other central regions (Diourbel, Fatick, and Louga) and between the accessions from Diourbel with those from Fatick and Louga. The lowest similarity values were observed between the accessions collected from the North (Saint-Louis) and the accessions from all other regions (similarity ranged from 0.861 with Thiès to 0.896 with Diourbel) (Table 3).
The genetic differentiation indices between populations (Fst) varied from 0.018 (between Thiès and the South) to 0.100 (between Thiès and Saint-Louis). The accessions collected in the southern part of the country showed a weak differentiation with the accessions from the other regions (0.018 ≤ Fst ≤ 0.064). The differentiations are low between accessions from this region and those from Fatick (Fst = 0.041), Louga (Fst = 0.031), Thiès (Fst = 0.018), and Diourbel (Fst = 0.021) regions but moderate between those from Saint-Louis (Fst = 0.064). The accessions collected in Saint-Louis showed the highest differentiations with the accessions from the other regions (0.085 ≤ Fst ≤ 0.100) (Table 4). All the pairwise Fst p values are significant (p value < 0.05).
Using the matrix of distances for genetic differentiations, an analysis of the molecular variance (AMOVA) was performed. The overall genetic variation has been divided among regions (11%), among individuals within regions (75%), and within individuals (14%). The results obtained showed that the diversity within regions (intra-regional diversity) was greater than the diversity between regions (inter-regional diversity). The observed Fst value was 0.114, suggesting moderate differentiation (Table 5).
The dendrogram shows three groups and several subgroups. Group I has the lowest number of accessions, collected from all regions except Fatick. Group II encompasses the majority of accessions into two large subgroups. The first is subdivided into a very heterogeneous subgroup containing accessions from all collection areas and a more or less homogeneous one containing most accessions from Saint-Louis (recession accessions grown in the post-rainy season) and some from Fatick. The second contains most accessions from Thiès and the southern part of Senegal. In the third group (III), accessions from all regions are also represented (Fig. 2).
The analysis of the population structure based to the method of Evanno et al. (Evanno et al. 2005) showed three populations (I, II, and III). Using a likelihood threshold of 0.55, 279 accessions (41.58%) mainly from Diourbel, Fatick, Thiès, and Sédhiou were assigned to population I, 218 accessions (23.49%) mainly from Louga and Diourbel to population II, and 116 accessions (17.28%) mainly from Saint-Louis (grown in the post-rainy season) to population III. The accessions which have likelihood thresholds lower than 0.55 were considered as an admixture. They represent 8.64% of the collection and consist of 58 accessions from all regions except the ones from Kolda and displayed a wide range of seed color. This group contains nearly half of the accessions from Sédhiou and some accessions from Fatick, Louga, Thiès, Diourbel, Saint-Louis, and Kédougou regions. Population II includes accessions with red-colored seed (58%) and the others, and population III the white-colored seeds (52.3%). Population I, which appears to be the least homogeneous, contains 20.3% of red-colored seed, 32.6% of white-colored seed, and 44.6% of black-colored seed. However, in each group, all the different colors and many regions are represented (Fig. 3).
D.ΔK criterion according to the calculation method by Evanno et al. (2005) (a) and structure of the Senegalese cowpea cultivated collection based on 15 SSR markers (b)
Genetic Diversity of Cowpea Wild Relatives
The wild relative samples consisted of two groups according to their type. Twenty-one (21) accessions are intermediate forms (weedy) including 20 from the Senegalese Midwest region (Louga, Fatick, and Diourbel) and 1 from the South (Kolda), and 45 are Vigna unguiculata var. spontanea from the Midwest (Thiès, Fatick, and Diourbel). These two groups (Vigna unguiculata var. spontanea and weedy were considered to investigate their level of genetic differentiation and variability. The Fst values detected between the wild relatives and the weedy were high and equal to 0.214 (Table 6) with a p value equal to 0.009 and a number of migrant per generation (Nm) equal to 0.623.
The cowpea wild relatives and intermediates structuring with DARwin revealed three groups. The first includes only one accession form Thiès. The second contained most of the accessions (49) from all the five collect regions. This group is subdivided into two subgroups. The first included 25 wild relative accessions from Thiès and Fatick (Vigna unguiculata var. spontanea) and the only weedy accession from Kolda. The second contained three wilds (2 from Fatick and 1 from Thiès), and the 20 accessions were identified as intermediate (weedy) forms. These types of cowpeas belong to the species Vigna unguiculata, often with spontaneous germination, and are only found in Louga, Diourbel, and Fatick regions. The third group contained 16 wild relative accessions from Thiès, Fatick, and Diourbel (Fig. 4).
The analysis of the wild relatives and intermediates (weedy) showed a best structure of the population at K equal 2 (Fig. 5a). Two groups were observed with group (i) encompassed all the weedy accessions (20 from Dioubel, Louga, and Fatick regions) except the one from the South (Kolda region). Group (ii) with 46 accessions included all the 45 wild relative accessions (from Thiès, Diourbel, Fatick) and the weedy one from Kolda (Fig. 5b). This clustering supported the structuration obtained with DARwin analysis, clearly showing the subdivision of wild/weedy into three genetic groups (Fig. 5c).
D.ΔK criterion according to the calculation method by Evanno et al. (2005) (a), clustering structure of the Senegalese cowpea wild relative and intermediate (weedy) accessions based on 15 SSR markers by structure (b) and Dwin (c)
Genetic Diversity and Structuration of Cultivated and Wild Relatives of Cowpea in Senegalese
The mean of the allele number (Na) was 6.2 for the cultivated accessions and 5.8 for the wild relatives. The values of the genetic diversity (mean expected heterozygosity (mHe)) and the fixation index (F), calculated for the two populations and based on the allelic frequencies for the 15 loci used, were higher for the wild relatives and weedy than for the cultivated forms. The differentiation index value (Fst) between the two groups was 0.104 with the number of migrants (Nm) being 3.086 (Table 7).
The structuring of all Senegalese cultivated, wild relative, and weedy accessions distinguishes local germplasm into two genetic groups (I and II) (Fig. 6a). The first one contained most of the cultivated accessions (573) including all those from Louga and Diourbel (147 and 158 respectively), 78 from Thiès, 78 from Fatick, 72 from Sédhiou, 26 from Saint-Louis, and 14 from Kédougou. The second group (with 154 accessions) gathered together all the wild relatives (Vigna unguiculata var. spontanea) and the intermediates called Kodj, and 92 cultivated accessions were collected from Fatick (4), Kolda (6), Kédougou (1), and Saint-Louis (81). This group contained the majority (81) of the accessions from Saint-Louis. Only four accessions (0.78%) from Fatick (2), Thiès (1), and Kédougou (1) showed values of probability of membership lower than 0.55 (shared among the two genetic populations) and were therefore classified as admixtures (Fig. 6b).
Population structure analysis for 737 cowpea accessions including 671 cultivated and 66 wild relatives and weedy based on microsatellite data. a Estimation of population using lnP(D) derived delta K with cluster number (K) ranged from 1 to10. b Two estimated populations (I and II) and five sub-populations (i, ii, iii, iv, and v) of Senegalese germplasm presented with different colors inferred by STRUCTURE analysis
In order to better understand the basis of the genetic structuration, the main populations (I and II) were further subdivided into five subpopulations (i–v) according to STRUCTURE with k = 5 the highest after k = 2 (Fig. 6a). It was observed that population I was subdivided into three subpopulations or subgroups (i–iii) and II into two (iv and v). In subgroups i, ii, and iii, accessions from all collection areas are present except those from Kolda region. Group iv contained all 45 wild relatives, 19 intermediate forms, and 7 cultivated accessions including 5 from Kolda and 2 from Thiès. The subgroup v, most homogeneous, consisted of 85 accessions from Saint-Louis (representing 90.22% of the subpopulation), 8 from Fatick, and 1 from Diourbel. Fifty-three (53) accessions were admixed from all regions of the study (Fig. 6b).
These data support the lack of clustering of accessions based on their geographical origin and the particularity of accessions from Saint-Louis region.
The dendrogram of genetic dissimilarities of the entire cultivated, wild relatives, and weedy (Kodj) cowpea collection also showed clearly five subpopulations (Fig. 7). The results supported the separation of the cowpea germplasm into five groups, which was also consistent with the model-based population structure. In summary, the model-based ancestry analysis, the phylogenetic tree and the principal coordinate analysis (PCoA) strongly supported that cowpea collection had five well-differentiated genetic populations and admixtures.
Discussion
Assessment of the genetic diversity of cowpea germplasm is a prerequisite for implementing a breeding program. The genetic diversity of cowpea accessions grown in studied locations in Senegal (Louga, Diourbel, Fatick, Thiès, Saint-Louis, Sédhiou, Kédougou, and Kolda) revealed a number of alleles ranging from 2 to 15 per locus. This number is relatively low and equivalent to those reported in Senegal by Badiane et al. (2012) (between 1 and 16) and in the germplasm from East Africa and IITA inbred lines by Desalegne et al. (2017) (between 4 and 15), but higher than the number of alleles reported by Li et al. (2001) (between 2 and 7) and Diouf and Hilu (2005) (between 1 and 9). This legume seems to be less diversified in Burkina Faso with the number of alleles ranging from 3 to 10, as reported by Sawadogo et al. (2010), and in Ethiopia from 2 to 5 by Desalegne et al. (2016). Similar results were obtained by Ogunkanmi et al. (2008) (number of alleles between 4 and 13) in a study based on 48 lines from different locations in Africa. The differences in the number of alleles reported by these different authors could be due to the type of material, the technique used for DNA separation during electrophoresis, allele detection, or the number of markers used in each study.
The values of the expected heterozygosity (between 0.317 and 0.439) in cowpea cultivated suggest low genetic diversity. These results were in agreement with previous studies on Senegalese cowpea diversity (Diouf and Hilu 2005; Kouadio et al. 2007; Badiane et al. 2012). This is attributed to the fact that cowpea is a self-pollinating crop, a bottleneck effect, or even to an evolutionary factor (natural selection) in favor of certain genotypes (Padulosi and Ng 1997). Nevertheless, the greatest cowpea diversity in this study was observed in southern Senegal in Sédhiou, Kolda, and Kédougou (0.439) regions and the lowest diversity in Fatick (0.317) in the center of Senegal. In the South, cowpea farmers’ ethnic groups are more diverse and their interest to different types of accessions might explain the diversity of the crop in this region (Labeyrie et al. 2014). In this part of the country, landraces are grown instead of improved varieties because the traits targeted in the national programs are more focused on the main constraints met in the Center and Center-North (Thiès, Louga, Diourbel, and Saint-Louis). In these latter areas, agriculture has been affected by a recurrent decrease in rainfall in recent decades. Indeed, the intensification of cowpea cultivation with the massive use of improved varieties adapted to these conditions explains the low diversity encountered. Furthermore, the analysis of the molecular variance (AMOVA) showed that, from one region to another, the genetic diversity was 11% while it was 75% among individuals within regions, and 14% within accessions. This very important intra-regional diversity could be linked to the presence of many different accessions in each region, while the low genetic diversity between regions could be partly explained by the distribution of the same cowpea seed (same varieties are found everywhere) in all the regions through donations, seed companies, or agricultural extension services. Accessions from the South seem genetically closest to those of all the other regions. These results were confirmed by the differentiation indices (Fst) that showed low differences between southern accessions compared with those from the other regions. This high genetic similarity could be explained by the fact that residents of some villages come from the central regions and have brought seed of some accessions with them according to our surveys (unpublished result). On the other hand, some accessions collected in this region are similar (in seed size and color) to those obtained in other regions and qualified as old varieties grown by the grandparents.
The dendrogram showed that only accessions from Thiès and those cultivated in flood recession soils in Saint-Louis showed a clear grouping, whereas those from other regions were less structured (dispersed in all three groups). Saint-Louis recession accessions have been well clustered, most likely due to time of cultivation. This type of cultivation is done after the raining season between November and February and is only practiced in this region. The difference between the periods of cultivation would limit gene flow, thus justifying the genetic differentiation noted. The presence of accessions from Louga and Diourbel in all groups is explained by the fact that these regions are located in the main cowpea production zone and the seeds are multiplied in Louga before their dissemination to the other regions by relevant services. Despite the low geographical structuring due to seed distribution across the country revealed by DARwin analysis, the grouping made without a priori with STRUCTURE software confirms the particularity of the accessions from Saint-Louis, which constitute the majority of population III. The lack of cowpea diversity structuring within a country is in line with the results of Desalegne et al. (2017) who analyzed cowpea genetic diversity in Ethiopia.
The percentage of hybrids detected in this study suggests a high rate of hybridization or gene flow in cowpea grown in Senegal. The gene flows in crop populations greatly depend on the exchange of seed, which is facilitated by social relationships (Leclerc and Coppens d’Eeckenbrugge 2011). In cowpea, hybridization percentages ranged from 1 to 9.5% (Kouam et al. 2012). These gene flows appear mainly as a constraint, limiting the differentiation between the varieties grown in the same area and the implementation of local adaptations (Ronfort et al. 2005; Kuruma et al. 2008).
The genetic differentiation indices noted between the wild relatives and the intermediate forms were high and significant (p value = 0.009). They showed important differentiations between Vigna unguiculata var. spontanea and weedy accessions. The number of migrants between these two groups was low (Nm = 0.623). These results suggested a low rate of genetic mixing between Vigna unguiculata var. spontanea from the center and the weedy.
The structuring carried out with DARwin and confirmed by STRUCTURE showed the genetic relationship between the wild from the center and the intermediate forms. These accessions have been grouped according to their type (wild relatives or intermediates). Some accessions of Vigna unguiculata var. spontanea and the weedy were included in a large group but were divided into two subgroups according to the type of accession. They should be evolved forms or cultivated introgressing wild alleles. This makes it possible to hypothesize that intermediates are semi-domesticated forms of wild, or are hybrids between wild and cultivated forms. This grouping supports the previous findings suggesting that V. unguiculata var. spontanea is the likely a progenitor of the domesticated cowpea (Padulosi and Ng 1997; Pasquet 1999).
This genetic diversity assessment study, using microsatellite markers, revealed slightly higher genetic diversity and fixation index in wild relatives compared with the cultivated accessions. Since its domestication, cowpea has maintained an evolutionary dynamic from the different growing areas. This dynamic is fuelled either with gene flow or natural selection pressure (Kouadio et al. 2007). However, the wild relatives remain more diverse than the cultivated forms (Ba et al. 2004; Ogunkanmi et al. 2008) because the latter have not been selected by humans according to specific attributes. The differences detected between wild/weedy and cultivated cowpea depend not only on the genetic variations of the accessions but also on a wider genetic base in the wild populations. A single microsatellite primer (VM 36) detected 13 alleles in 48 wild-type (Ogunkanmi et al. 2008) accessions, while it detected only 7 in 91 cowpea selection lines (Li et al. 2001). The observed differentiation index value (Fst = 0.104) suggested moderate differentiation between Senegalese wild/weedy and cultivated cowpea. Our results also indicate a gene flow (Nm = 3.086) between these two groups, suggesting that it could be due to gene exchange between cowpea cultivated and wild relative species. Similar results were reported in Ethiopian cowpea germplasm, with a differentiation index and a number of migrants per generation equal to 0.075 and 3.176, respectively (Desalegne et al. 2016). The structure analysis revealed two genetic populations within Senegalese cowpea cultivated and wild/weedy germplasm. Population I included only cultivated accessions, while population II gathered together all the wild relatives (Vigna unguiculata var. spontanea) and the intermediates, and some cultivated accessions of which the majority were collected from the Northern (Saint-Louis region). Therefore, Senegalese wild populations did not give rise to all cultivated forms collected. These two main populations were divided into five subgroups (i–v) for a better understanding of genetic basis. This structure clustering, well-supported by both DARwin and PCoA analyses, showed a discrimination of cultivated and wild/weedy populations. The separation of wild and domesticated cowpea gene pools observed with SSR data is in agreement with isozyme and AFLP (Pasquet 1999; Coulibaly et al. 2002) and RAPD (Ba et al. 2004) data. In neither case was any evidence found to support clustering of accessions based on their collection areas like in Ethiopian (Desalegne et al. 2016). Regardless of the region of origin of the samples, no grouping of wild accessions with those cultivated in the same region was observed. This suggested gene flows between wild relatives rather than between wild and cultivated from the same region.
Conclusion
The genetic variability and population structure of Senegalese cowpea germplasm including cultivated, wild relatives, and weedy accessions were assessed in this study. The study showed that the genetic structure does not depend on regions. The difference between cultivated and wild/weedy accessions in genetic structure indicated that there are still abundant genetic resources in local germplasm, which show good adaptation in local areas, especially under biotic and abiotic stress environments. The wild relative genotypes are significantly differentiated from those cultivated in Senegal, suggesting that they could provide new genes for the improvement of local germplasm. The results obtained from this study will allow a better use and exploitation of Senegalese germplasm in breeding programs and better conservation.
References
Agbicodo EM, Fatokun CA, Muranaka S, Visser RGF, van der CG L (2009) Breeding drought tolerant cowpea: constraints, accomplishments, and future prospects. Euphytica 167:353–370. https://doi.org/10.1007/s10681-009-9893-8
Agence Nationale de la Statistique et de la Démographie (ANSD) (2019) Bulletin mensuel des statistiques économiques de Mai 2019. In: ansd.sn. http://www.ansd.sn/
Asare AT, Gowda BS, Galyuon IKA, Aboagye LL, Takrama JF, Timko MP (2010) Assessment of the genetic diversity in cowpea ( Vigna unguiculata L. Walp.) germplasm from Ghana using simple sequence repeat markers. Plant Genet Resour 8:142–150. https://doi.org/10.1017/S1479262110000092
Ba FS, Pasquet RS, Gepts P (2004) Genetic diversity in cowpea [Vigna unguiculata (L.) Walp.] as revealed by RAPD markers. Genet Resour Crop Evol 51:539–550. https://doi.org/10.1023/B:GRES.0000024158.83190.4e
Badiane FA, Gowda BS, Cissé N, Diouf D, Sadio O, Timko MP (2012) Genetic relationship of cowpea (Vigna unguiculata) varieties from Senegal based on SSR markers. Genet Mol Res 11:292–304. https://doi.org/10.4238/2012.February.8.4
Badiane FA, Diouf M, Diouf D (2014) Cowpea. In: Singh M, Bisht IS, Dutta M (eds) Broadening the genetic base of grain legumes. Springer India, New Delhi, pp 95–114
Barrett BA, Kidwell KK (1998) AFLP-based genetic diversity assessment among wheat cultivars from the Pacific Northwest. Crop Sci 38:1261–1271. https://doi.org/10.2135/cropsci1998.0011183X003800050025x
Boukar O, Massawe F, Muranaka S, Franco J, Maziya-Dixon B, Singh B, Fatokun C (2011) Evaluation of cowpea germplasm lines for protein and mineral concentrations in grains. Plant Genet Resour 9:515–522. https://doi.org/10.1017/S1479262111000815
Cisse N, Hall AE (2003) Traditional cowpea in Senegal, a case study. www.fao.org/ag/AGP/AGPC/doc/publicat/cowpea_Cisse/cowpea_cisse_e.htm[2007]
Coulibaly S, Pasquet RS, Papa R, Gepts P (2002) AFLP analysis of the phenetic organization and genetic diversity of Vigna unguiculata L. Walp. reveals extensive gene flow between wild and domesticated types. Theor Appl Genet 104:358–366. https://doi.org/10.1007/s001220100740
Desalegne BA, Mohammed S, Dagne K, Timko MP (2016) Assessment of genetic diversity in Ethiopian cowpea [Vigna unguiculata (L.) Walp.] germplasm using simple sequence repeat markers. Plant Mol Biol Report 34:978–992. https://doi.org/10.1007/s11105-016-0979-x
Desalegne BA, Dagne K, Melaku G, Ousmane B, Fatokun CA (2017) Efficiency of SNP and SSR-based analysis of genetic diversity, population structure, and relationships among cowpea (Vigna unguiculata (L.) Walp.) germplasm from East Africa and IITA inbred lines. J Crop Sci Biotechnol 20:107–128. https://doi.org/10.1007/s12892-016-0051-0
Dikshit HK, Jhang T, Singh NK, Koundal KR, Bansal KC, Chandra N, Tickoo JL, Sharma TR (2007) Genetic differentiation of Vigna species by RAPD, URP and SSR markers. Biol Plant 51:451–457. https://doi.org/10.1007/s10535-007-0095-8
Diouf D (2011) Recent advances in cowpea [Vigna unguiculata (L.) Walp.] omics research for genetic improvement. Afr J Biotechnol 10:2803–2819. https://doi.org/10.5897/AJBx10.015
Diouf D, Hilu KW (2005) Microsatellites and RAPD markers to study genetic relationships among cowpea breeding lines and local varieties in Senegal. Genet Resour Crop Evol 52:1057–1067. https://doi.org/10.1007/s10722-004-6107-z
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Egbadzor KF, Ofori K, Yeboah M, Aboagye LM, Opoku-Agyeman MO, Danquah EY, Offei SK (2014) Diversity in 113 cowpea [Vigna unguiculata (L) Walp] accessions assessed with 458 SNP markers. SpringerPlus 3: . https://doi.org/10.1186/2193-1801-3-541
Ehlers JD, Hall AE (1997) Cowpea (Vigna unguiculata L. Walp.). Field Crops Res 53:187–204. https://doi.org/10.1016/S0378-4290(97)00031-2
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
FAOSTAT (2014) Agricultural production. Crop primary database. Food and Agricultural Organization of the United Nations. In: fao.org. http://faostat.fao.org/faostat
Fatokun CA, Danesh D, Young ND, Stewart EL (1993) Molecular taxonomic relationships in the genus Vigna based on RFLP analysis. Theor Appl Genet 86:97–104. https://doi.org/10.1007/BF00223813
Fotso M, Azanza J-L, Pasquet R, Raymond J (1994) Molecular heterogeneity of cowpea (Vigna unguiculata Fabaceae) seed storage proteins. Plant Syst Evol 191:39–56. https://doi.org/10.1007/BF00985341
Ghalmi N, Malice M, Jacquemin J-M, Ounane S-M, Mekliche L, Baudoin J-P (2010) Morphological and molecular diversity within Algerian cowpea (Vigna unguiculata (L.) Walp.) landraces. Genet Resour Crop Evol 57:371–386. https://doi.org/10.1007/s10722-009-9476-5
Gupta SK, Gopalakrishna T (2010) Development of unigene-derived SSR markers in cowpea ( Vigna unguiculata ) and their transferability to other Vigna species. Genome 53:508–523. https://doi.org/10.1139/G10-028
Hall AE (2004) Breeding for adaptation to drought and heat in cowpea. Eur J Agron 21:447–454. https://doi.org/10.1016/j.eja.2004.07.005
Hall AE, Cisse N, Thiaw S, Elawad HOA, Ehlers JD, Ismail AM, Fery RL, Roberts PA, Kitch LW, Murdock LL, Boukar O, Phillips RD, McWatters KH (2003) Development of cowpea cultivars and germplasm by the bean/cowpea CRSP. Field Crops Res 82:103–134. https://doi.org/10.1016/S0378-4290(03)00033-9
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Kouadio D, Echikh N, Toussaint A, Pasquet RS, Baudoin J-P (2007) Organisation du pool génique de Vigna unguiculata (L.) Walp.: croisements entre les formes sauvages et cultivées du niébé. Biotechnol Agron Soc Env 11
Kouakou CK, Roy-Macauley H, Coudou M, Otto MC, Rami J-F, Cissé N (2007) Diversité génétique des variétés traditionnelles de niébé [Vigna unguiculata (L.) Walp.] au Sénégal: étude préliminaire. Plant Genet Resour Newsl 152:33–44
Kouam EB, Pasquet RS, Campagne P, Tignegre J-B, Thoen K, Gaudin R, Ouedraogo JT, Salifu AB, Muluvi GM, Gepts P (2012) Genetic structure and mating system of wild cowpea populations in West Africa. BMC Plant Biol 12:113. https://doi.org/10.1186/1471-2229-12-113
Kuruma RW, Kiplagat O, Ateka E, Owuoche G (2008) Genetic diversity of Kenyan cowpea accessions based on morphological and microsatellite markers. East Afr Agric For J 76:8
Labeyrie V, Deu M, Barnaud A, Calatayud C, Buiron M, Wambugu P, Manel S, Glaszmann J-C, Leclerc C (2014) Influence of ethnolinguistic diversity on the sorghum genetic patterns in subsistence farming systems in Eastern Kenya. PLoS One 9:e92178. https://doi.org/10.1371/journal.pone.0092178
Lal H, Rai N, Rai KK, Tiwari SK (2016) Microsatellites markers to study genetic relationships among cowpea (Vigna unguiculata) genotypes. Indian J Agri Sci 86:8
Leclerc C, Coppens d’Eeckenbrugge G (2011) Social organization of crop genetic diversity. The G × E × S interaction model. Diversity 4:1–32. https://doi.org/10.3390/d4010001
Li Wang M, Barkley NA, Gillaspie GA, Pederson GA (2008) Phylogenetic relationships and genetic diversity of the USDA Vigna germplasm collection revealed by gene-derived markers and sequencing. Genet Res 90:467–480. https://doi.org/10.1017/S0016672308009889
Li C-D, Fatokun CA, Ubi B, Singh BB, Scoles GJ (2001) Determining genetic similarities and relationships among cowpea breeding lines and cultivars by microsatellite markers. Crop Sci 41:189–197. https://doi.org/10.2135/cropsci2001.411189x
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. 89:583–590
Nkongolo KK (2003) Genetic characterization of Malawian cowpea (Vigna unguiculata (L.) Walp) landraces: diversity and gene flow among accessions. Euphytica 129:219–228. https://doi.org/10.1023/A:1021935431873
O’Neill R, Snowdon R, Köhler W (2003) Population genetics: aspects of biodiversity. In: Esser K, Lüttge U, Beyschlag W, Hellwig F (eds) Progress in botany. Springer, Berlin, pp 115–137
Obilana AT (1987) Breeding cowpeas for Striga resistance, parasitic weed in agriculture. Musselman L.J, CRC Press. Boca Raton
Ogunkanmi LA, Ogundipe OT, Ng NQ, Fatokun CA (2008) Genetic diversity in wild relatives of cowpea (Vigna unguiculata) as revealed by simple sequence repeats (SSR) markers. J Food Agric Environ 6:263–268
Padulosi S, Ng NQ (1997) Origin, taxonomy, and morphology of Vigna unguiculata (L.) Walp., Advances_in_Cowpea_Research. Singh B.B., MohanRaj D.R., Dashiell K.E. and Jackai L.E.N., IITA-JIRCAS, Ibadan
Panella L, Gepts P (1992) Genetic relationships within Vigna unguiculata (L) Walp based on isozyme analyses. Genet Resour Crop Evol 39:71–88. https://doi.org/10.1007/BF00051226
Pasquet RS (1993) Variation at isozyme loci in wild Vigna unguiculata (Fabaceae, Phaseoleae). Plant Syst Evol 186:157–173. https://doi.org/10.1007/BF00940795
Pasquet RS (1999) Genetic relationships among subspecies of Vigna unguiculata (L.) Walp. based on allozyme variation. Theor Appl Genet 98:1104–1119. https://doi.org/10.1007/s001220051174
Pasquet RS, Baudoin JP (1997) Cowpea. In: Tropical plant breeding. Charrier A., Jacquot M., Hamon S., Nicolas D., CIRAD, pp 177–198
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539. https://doi.org/10.1093/bioinformatics/bts460
Perrier, X, Jacquemoud-Collet, JP (2016) DARwin Software
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Risterucci AM, Grivet L, N’Goran JAK, Pieretti I, Flament MH, Lanaud C (2000) A high-density linkage map of Theobroma cacao L. Theor Appl Genet 101:948–955. https://doi.org/10.1007/s001220051566
Ronfort J, Jenczewski E, Muller MH (2005) Les flux de gènes et leur impact sur la structure de la diversité génétique. Le cas des prairies Fourrages 182:275–286
Sawadogo M, Ouedraogo JT, Gowda BS, Timko MP (2010) Genetic diversity of cowpea (Vigna unguiculata L. Walp.) cultivars in Burkina Faso resistant to Striga gesnerioides. Afr J Biotechnol 9:8146–8153
Séne D (1966) Inventaire des principales variétés de niébé (Vigna unguiculata Walpers) cultivées au Sénégal. L’Agronomie Trop:927–933
Simon MV, Benko-Iseppon A-M, Resende LV, Winter P, Kahl G (2007) Genetic diversity and phylogenetic relationships in Vigna Savi germplasm revealed by DNA amplification fingerprinting. Genome 50:538–547. https://doi.org/10.1139/G07-029
Singh BB (2002) Breeding cowpea varieties for resistance to Striga gesnerioides and Alectravogelii., challenges and opportunities for enhancing sustainable cowpea production: proceedings of the World Cowpea Conference III held at IITA, 4–8 September 2000. Fatokun C.A., et al., Ibadan. Nigeria
Singh BB, Sharma B (1996) Restructuring cowpea for higher yield. Indian J Genet Plant Breed 56:389–405
Spencer M, Ndiaye M, Gueye M, Diouf D, Ndiaye MA (2000) DNA-based relatedness of cowpea [Vigna unguiculata (L.) Walp.] genotypes using DNA amplification fingerprinting—UQ eSpace. Physiol. Mol. Biol. Plants 81–88
Uma MS, Hittalamani S, Murthy BCK, Viswanatha KP (2009) Microsatellite DNA marker aided diversity analysis in cowpea [Vigna unguiculata (L.) Walp]. Indian J Genet Plant Breed 69:35–43
Vaillancourt RE, Weeden NF (1992) Chloroplast DNA polymorphism suggests Nigerian center of domestication for the cowpea, Vigna unguiculata (Leguminosae). Am J Bot 79:1194–1199. https://doi.org/10.1002/j.1537-2197.1992.tb13716.x
Xu P, Wu X, Wang B, Liu Y, Qin D, Ehlers JD, Close TJ, Hu T, Lu Z, Li G (2010) Development and polymorphism of Vigna unguiculata ssp. unguiculata microsatellite markers used for phylogenetic analysis in asparagus bean (Vigna unguiculata ssp. sesquipedialis (L.) Verdc.). Mol Breed 25:675–684. https://doi.org/10.1007/s11032-009-9364-x
Acknowledgments
We thank the staff of “Direction Régional du Développement Rural (DRDR)” who guided and assisted us during the collection of the plant materials, Mr. Mbaye Ndoye Sall for his support for the laboratory experiments, Rémy S. Pasquet and Abdoulaye NGom for their help for identifying cowpea wild relative species and intermediate forms, and Katina Olodo and Oberline Fokou for their helpful suggestions. We are also very grateful to the anonymous reviewers for their helpful comments.
Availability of Data and Materials
Data and materials are available.
Code Availability
Not applicable.
Funding
The authors would like to thank the West African Agricultural Productivity Program (WAAPP) for the financial support.
Author information
Authors and Affiliations
Contributions
Awa SARR, Amy BODIAN, and Kodjo Mawuena Gbedevi collected the plant material; Awa SARR, Amy BODIAN, and Diaga DIOUF conceived and designed the experiments; Awa SARR, Kodjo Mawuena Gbedevi, and Elisabeth DIOP performed the DNA genotyping; Awa SARR, Amy BODIAN, and Baye Maguette DIOP analyzed the data; Awa SARR drafted the paper; and Amy BODIAN, Daniel Fonceka, Diaga DIOUF, Oyatomi Olaniyi A., Badara GUEYE, Ndiaga CISSÉ, Baye Maguette DIOP, and Khadidiatou Ndoye NDIR edited and provided critical review of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest.
Ethics Approval and Consent to Participation
Not applicable.
Consent for Publication
Not applicable
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key Message
This study including the cultivated cowpea and wild relative accessions allowed to characterize a large collection representative of cowpea local germplasm. It was undertaken to address the lack of knowledge about Senegalese cowpea resources to improve the effectiveness of breeding programs and guide future germplasm conservation efforts.
Electronic Supplementary Material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sarr, A., Bodian, A., Gbedevi, K.M. et al. Genetic Diversity and Population Structure Analyses of Wild Relatives and Cultivated Cowpea (Vigna unguiculata (L.) Walp.) from Senegal Using Simple Sequence Repeat Markers. Plant Mol Biol Rep 39, 112–124 (2021). https://doi.org/10.1007/s11105-020-01232-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-020-01232-z