Abstract
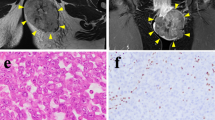
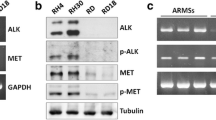
Alveolar rhabdomyosarcoma (aRMS) is a histological subtype of RMS, which is the most common pediatric and adolescent soft-tissue sarcoma, accounting for 3–4% of all pediatric malignancies. Patient-derived cells are essential tools for understanding the molecular mechanisms of poor prognosis and developing novel anti-cancer drugs. However, only a limited number of well-characterized cell lines for rhabdomyosarcoma from public cell banks is available. Therefore, we aimed to establish a novel cell line of aRMS from the tumor tissue of a patient with aRMS. The cell line was established from surgically resected tumor tissue from a 4-year-old male patient diagnosed with stage III, T2bN1M0 aRMS and was named as NCC-aRMS1-C1. The cells were maintained for more than 3 months under tissue culture conditions and passaged more than 20 times. We confirmed the presence of identical fusion gene such as PAX7-FOXO1 in both the original tumor and NCC-aRMS1-C1. The cells exhibited spheroid formation and invasion. We found that docetaxel, vincristine, ifosfamide, dacarbazine, and romidepsin showed remarkable growth-suppressive effects on the NCC-aRMS1-C1 cells. In conclusion, the NCC-aRMS1-C1 cell line exhibited characteristics that may correspond to the lymph node metastasis in aRMS and mirror its less aggressive features. Thus, it may be useful for innovative seeds for novel therapeutic strategies.





Similar content being viewed by others
References
Amer KM, Thomson JE, Congiusta D, et al. Epidemiology, incidence, and survival of rhabdomyosarcoma subtypes: SEER and ICES database analysis. J Orthopaedic Res. 2019;37:2226–30.
Parham DM, Barr FG. Classification of rhabdomyosarcoma and its molecular basis. Adv Anat Pathol. 2013;20:387–97.
Matsumura T, Yamaguchi T, Seki K, et al. Advantage of FISH analysis using FKHR probes for an adjunct to diagnosis of rhabdomyosarcomas. Virchows Arch. 2008;452:251–8.
Skapek SX, Anderson J, Barr FG, et al. PAX-FOXO1 fusion status drives unfavorable outcome for children with rhabdomyosarcoma: a children's oncology group report. Pediatr Blood Cancer. 2013;60:1411–7.
Duan F, Smith LM, Gustafson DM, et al. Genomic and clinical analysis of fusion gene amplification in rhabdomyosarcoma: a report from the Children's Oncology Group. Genes Chromosomes Cancer. 2012;51:662–74.
Missiaglia E, Williamson D, Chisholm J, et al. PAX3/FOXO1 fusion gene status is the key prognostic molecular marker in rhabdomyosarcoma and significantly improves current risk stratification. J Clin Oncol. 2012;30:1670–7.
Bridge JA, Liu J, Qualman SJ, et al. Genomic gains and losses are similar in genetic and histologic subsets of rhabdomyosarcoma, whereas amplification predominates in embryonal with anaplasia and alveolar subtypes. Genes Chromosomes Cancer. 2002;33:310–21.
Kephart JJ, Tiller RG, Crose LE, et al. Secreted Frizzled-Related Protein 3 (SFRP3) is required for tumorigenesis of PAX3-FOXO1-positive alveolar rhabdomyosarcoma. Clin Cancer Res. 2015;21:4868–80.
Saab R, Spunt SL, Skapek SX. Myogenesis and rhabdomyosarcoma the Jekyll and Hyde of skeletal muscle. Curr Top Dev Biol. 2011;94:197–234.
Maurer HM, Gehan EA, Beltangady M, et al. The intergroup rhabdomyosarcoma study-II. Cancer. 1993;71:1904–22.
Maurer HM, Beltangady M, Gehan EA, et al. The intergroup rhabdomyosarcoma study-I. A final report. Cancer. 1988;61:209–20.
Koscielniak E, Harms D, Henze G, et al. Results of treatment for soft tissue sarcoma in childhood and adolescence: a final report of the German Cooperative Soft Tissue Sarcoma Study CWS-86. J Clin Oncol. 1999;17:3706–19.
Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer. 2010;10:241–53.
Barretina J, Caponigro G, Stransky N, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7.
Garnett MJ, Edelman EJ, Heidorn SJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5.
Basu A, Bodycombe NE, Cheah JH, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154:1151–61.
Seashore-Ludlow B, Rees MG, Cheah JH, et al. Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov. 2015;5:1210–23.
Rees MG, Seashore-Ludlow B, Cheah JH, et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol. 2016;12:109–16.
Haverty PM, Lin E, Tan J, et al. Reproducible pharmacogenomic profiling of cancer cell line panels. Nature. 2016;533:333–7.
Iorio F, Knijnenburg TA, Vis DJ, et al. A landscape of pharmacogenomic interactions in cancer. Cell. 2016;166:740–54.
Behan FM, Iorio F, Picco G, et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature. 2019;568:511–6.
Raney RB, Walterhouse DO, Meza JL, et al. Results of the Intergroup Rhabdomyosarcoma Study Group D9602 protocol, using vincristine and dactinomycin with or without cyclophosphamide and radiation therapy, for newly diagnosed patients with low-risk embryonal rhabdomyosarcoma: a report from the Soft Tissue Sarcoma Committee of the Children's Oncology Group. J Clin Oncol. 2011;29:1312–8.
Barr FG, Nauta LE, Davis RJ, Schäfer BW, Nycum LM, Biegel JA. In vivo amplification of the PAX3-FKHR and PAX7-FKHR fusion genes in alveolar rhabdomyosarcoma. Hum Mol Genet. 1996;5:15–211.
Reichek JL, Duan F, Smith LM, et al. Genomic and clinical analysis of amplification of the 13q31 chromosomal region in alveolar rhabdomyosarcoma: a report from the Children's Oncology Group. Clin Cancer Res. 2011;17:1463–73.
Yoshimatsu Y, Noguchi R, Tsuchiya R, et al. Establishment and characterization of NCC-CDS2-C1: a novel patient-derived cell line of CIC-DUX4 sarcoma. Hum Cell. 2020;33:427–36.
Bairoch A. The Cellosaurus, a cell-Line knowledge resource. J Biomol Tech. 2018;29:25–38.
Billiau A, Edy VG, Heremans H, et al. Human interferon: mass production in a newly established cell line, MG-63. Antimicrob Agents Chemother. 1977;12:11–5.
Sorensen PH, Lynch JC, Qualman SJ, et al. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children's oncology group. J Clin Oncol. 2002;20:2672–9.
Bennicelli JL, Advani S, Schäfer BW, Barr FG. PAX3 and PAX7 exhibit conserved cis-acting transcription repression domains and utilize a common gain of function mechanism in alveolar rhabdomyosarcoma. Oncogene. 1999;18:4348–56.
Leu KM, Ostruszka LJ, Shewach D, et al. Laboratory and clinical evidence of synergistic cytotoxicity of sequential treatment with gemcitabine followed by docetaxel in the treatment of sarcoma. J Clin Oncol. 2004;22:1706–12.
Bui BN, Chevallier B, Chevreau C, et al. Efficacy of lenograstim on hematologic tolerance to MAID chemotherapy in patients with advanced soft tissue sarcoma and consequences on treatment dose-intensity. J Clin Oncol. 1995;13:2629–36.
Nakajima H, Kim YB, Terano H, Yoshida M, Horinouchi S. FR901228, a potent antitumor antibiotic, is a novel histone deacetylase inhibitor. Exp Cell Res. 1998;241:126–33.
Barbarotta L, Hurley K. Romidepsin for the treatment of peripheral T-cell lymphoma. J Adv Pract Oncol. 2015;6:22–36.
Piekarz RL, Frye R, Turner M, et al. Phase II multi-institutional trial of the histone deacetylase inhibitor romidepsin as monotherapy for patients with cutaneous T-cell lymphoma. J Clin Oncol. 2009;27:5410–7.
Blattmann C, Oertel S, Ehemann V, et al. Enhancement of radiation response in osteosarcoma and rhabdomyosarcoma cell lines by histone deacetylase inhibition. Int J Radiat Oncol Biol Phys. 2010;78:237–45.
Kutko MC, Glick RD, Butler LM, et al. Histone deacetylase inhibitors induce growth suppression and cell death in human rhabdomyosarcoma in vitro. Clin Cancer Res. 2003;9:5749–55.
Vleeshouwer-Neumann T, Phelps M, Bammler TK, MacDonald JW, Jenkins I, Chen EY. Histone deacetylase inhibitors antagonize distinct pathways to suppress tumorigenesis of embryonal rhabdomyosarcoma. PLoS ONE. 2015;10:e0144320.
Hedrick E, Crose L, Linardic CM, Safe S. Histone deacetylase inhibitors inhibit rhabdomyosarcoma by reactive oxygen species-dependent targeting of specificity protein transcription factors. Mol Cancer Ther. 2015;14:2143–53.
Acknowledgements
We thank Drs. M. Endo, Y. Minami, K. Shimizu, T. Mori, T. Uehara M. Sugawara, Y. Araki, S. Toki, and Ms. R. Nakano from the Division of Musculoskeletal Oncology, National Cancer Center Hospital for sampling tumor tissue specimens from surgically resected materials. We appreciate the technical assistance of Ms. Y. Kuwata (Division of Rare Cancer Research, National Cancer Center Institute). We also appreciate the technical support provided by Ms Y. Shiotani, Mr. N. Uchiya, and Dr. T. Imai (Central Animal Division, National Cancer Center Research Institute). Lastly, we also thank Editage (www.editage.jp) for English language editing and constructive comments on the manuscript.
Funding
This research was supported by the Japan Agency for Medical Research and Development (Grant Number 20ck0106537h0001), “Study to Overcome the Limits of Cancer Genome-based Medicine Using Patient-derived “Rare Cancer” Model.” The authors thank all those who participated in the study and helped facilitate the research process.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Ethics approval and consent to participate
This study was approved by the ethics committee of the National Cancer Center.
Consent of publication
Written informed consent was obtained from the parents of the patient.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sin, Y., Yoshimatsu, Y., Noguchi, R. et al. Establishment and characterization of a novel alveolar rhabdomyosarcoma cell line, NCC-aRMS1-C1. Human Cell 33, 1311–1320 (2020). https://doi.org/10.1007/s13577-020-00403-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-020-00403-0