Abstract
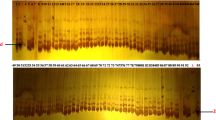
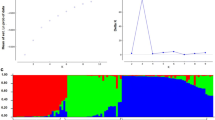
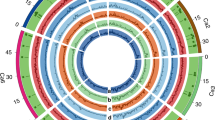
Chickpea (Cicer arietinum L.) is the third most important legume crop as a stable protein source in human feed. Breeding efforts in chickpea need to find the genotypes with diverse genome background for crossing to produce progenies that are used in the evaluation of favorable traits. In this study, we employed DArTseq-generated SilicoDArT markers for assessment of genetic diversity, population structure and linkage disequilibrium in a panel of 90 chickpea advanced breeding lines. Totally, 9824 SilicoDArT markers generated through DArTseq genotyping and, after filtering, 2053 markers with an average of 265.5 markers per chromosome were used for genetic diversity analysis. Polymorphism information content (PIC) value of SilicoDArT markers ranged from 0.05 to 0.50, with an average of 0.25. Extensive and low level of LD decay in long distances with average r2 = 0.15 was observed. Chickpea genotypes showed high genetic diversity with average kinship value and genetic distance of − 0.54 and 0.36, respectively. Results of cluster analysis, population structure and discriminant analysis of principal component (DAPC) were consistent together in grouping chickpea genotypes into four distinct clusters. These findings demonstrated the efficiency of SilicoDArT markers for large-scale diversity analysis in chickpea, and results can be used for future genomic studies in chickpea such as genome-wide association study and genomic selection for important traits such as seed yield and resistance to abiotic and biotic stresses.






Similar content being viewed by others
References
Abbo S, Berger J, Turner NC (2003) Viewpoint: evolution of cultivated chickpea: four bottlenecks limit diversity and constrain adaptation. Funct Plant Biol 30:1081–1087
Ahmad F, Khan AI, Awan FS, Sadia B, Sadaqat HA, Bahadur S (2010) Genetic diversity of chickpea (Cicer arietinum L.) germplasm in Pakistan as revealed by RAPD analysis. Genet Mol Res 9:1414–1420
Aslam M, Mahmood IA, Peoples MB, Schwenke GD, Herridge DF (2003) Contribution of chickpea nitrogen fixation to increased wheat production and soil organic fertility in rain-fed cropping. Biol Fertil Soils 38:59–64
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Dodds KG, McEwan JC, Brauning R, Anderson RA, Van Stijn TC, Kristjánsson T, Clarke S (2015) Construction of relatedness matrices using genotyping-by-sequencing data. BMC Genom 16:1047
Egea LA, Mérida-García R, Kilian A, Hernandez P, Dorado G (2017) Assessment of genetic diversity and structure of large garlic (Allium sativum) germplasm bank, by diversity arrays technology “genotyping-by-sequencing” platform (DArTseq). Front Genet 8:98
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Farahani S, Maleki M, Mehrabi R, Kanouni H, Scheben A, Batley J, Talebi R (2019) Whole genome diversity, population structure and linkage disequilibrium analysis of chickpea (Cicer arietinum L.) advanced breeding lines using genome-wide DArTseq-based SNP markers. Genes 10:676
FAOSTAT (2015) FAO statistical databases. Food and Agricultural Organization. https://faostat.fao.org/. Accessed 9 Nov 2017
Fayaz F, Aghaee M, Talebi R, Azadi A (2019) Genetic diversity and molecular characterization of iranian durum wheat landraces (Triticum turgidum durum (Desf.) Husn.) using DArT markers. Biochem Genet 57:98–116
Ghaffari P, Talebi R, Keshavarz F (2014) Genetic diversity and geographical differentiation of Iranian landrace, cultivars and exotic chickpea lines as revealed by morphological and microsatellite markers. Physiol Mol Biol Plant 20:225–233
Gupta S, Nawaz K, Parween S, Roy R, Sahu K, Pole AK, Khandal H, Srivastava R, Parida SK, Chattopadhyay D (2017) Draft genome sequence of Cicer reticulatum L., the wild progenitor of chickpea provides a resource for agronomic trait improvement. DNA Res 24:1–10
Hajibarat Z, Saidi A, Hajibarat Z, Talebi R (2014) Genetic diversity and population structure analysis of landrace and improved chickpea (Cicer arietinum) genotypes using morphological and microsatellite markers. Environ Exp Biol 12:161–166
Hajibarat Z, Saidi A, Hajibarat Z, Talebi R (2015) Characterization of genetic diversity in chickpea using SSR markers, start codon targeted polymorphism (SCoT) and conserved DNA-derived polymorphism (CDDP). Physiol Mol Biol Plant 21:365–373
Hassani SMR, Talebi R, Pourdad SS, Naji AM, Fayaz F (2020) Genetic diversity, population structure and linkage disequilibrium analysis of worldwide diverse safflower (Carthamus tinctorius L.) accessions using genotyping-by-sequencing (GBS) through DArTseq platform. Mole Biol Rep 47:2123–2135
Iruela M, Rubio J, Cubero JI, Gil J, Milan T (2002) Phylogenetic analysis in the genus Cicer and cultivated chickpea using RAPD and ISSR markers. Theor Appl Genet 104:643–651
Jain M, Misra G, Patel RK, Priya P, Jhanwar S, Khan AW, Shah N, Singh VK, Garg R, Jeena G, Yadav M, Kant C, Sharma P, Yadav G, Bhatia S, Tyagi AK, Chattopadhyay D (2013) A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant J 74:715–29
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24(11):1403–1405
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Khan MA, Akhtar N, Ullah I, Jaffery S (1995) Nutritional evaluation of desi and kabuli chickpeas and their products commonly consumed in Pakistan. Int J Food Sci Nutr 46:215–223
Kilian A, Huttner E, Wenzl P, Jaccoud D, Carling J, Caig V, Evers M, Heller-Uszynska K, Cayla C, Patarapuwadol S, Xia L (2003) The fast and the cheap: SNP and DArT-based whole genome profiling for crop improvement. In: Tuberosa R, Phillips RL, Gale M (eds) Proceedings of the international congress In the wake of the double helix: from the green revolution to the gene revolution, Bologna, pp 443–461
Kilian A, Wenzl P, Huttner E, Carling J, Xia L, Blois H, Caig V, Heller-Uszynsk K, Jaccoud D, Hopper C, Aschenbrenner-Kilian M, Evers M, Peng K, Cayla C, Hok P, Uszynski G (2012) Diversity arrays technology: a generic genome profiling technology on open platforms. Methods Mol Biol 888:67–89
Kujur A, Bajaj D, Upadhyaya HD, Das S, Ranjan R, Shree T, Saxena MS, Badoni S, Kumar V, Tripathi S, Gowda CLL, Sharma S, Singh S, Tyagi AK, Parida SK (2015) Employing genome-wide SNP discovery and genotyping strategy to extrapolate the natural allelic diversity and domestication patterns in chickpea. Front Plant Sci 6:162
Kumar J, van Rheenen HA (2000) A major gene for time of flowering in chickpea. J Hered 91:67–68
Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28:2397–2399
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Mahboubi M, Mehrabi R, Naji AM, Talebi R (2020) Whole-genome diversity, population structure and linkage disequilibrium analysis of globally diverse wheat genotypes using genotyping-by-sequencing DArTseq platform. 3Biotech 10:48
Monostori I, Szira F, Tondelli A, Arendas T, Gierczik K, Cattivelli L, Galiba G, Vagujfalvi A (2017) Genome-wide association study and genetic diversity analysis on nitrogen use efficiency in a Central European winter wheat (Triticum aestivum L.) collection. PloS One 2:e0189265
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 8:4321–4326
Nadeem MA, Habyarimana E, Ciftci V, Nawaz MA, Karakoy T, Comertpay G, Shahid MQ, Hatipoğlu R, Yeken MZ, Ali F, Ercişli S, Gyuhwa Chung G, Baloch FS (2018) Characterization of genetic diversity in Turkish common bean gene pool using phenotypic and whole-genome DArTseq-generated silicoDArT marker information. PLoS ONE 13:e0205363
O’Connor K, Kilian A, Hayes B, Hardner C, Nock C, Baten A, Alam M, Topp B (2019) Population structure, genetic diversity and linkage disequilibrium in a macadamia breeding population using SNP and silicoDArT markers. Tree Genet Genom 15:24
Pakseresht F, Talebi R, Karami E (2013) Comparative assessment of ISSR, DAMD and SCoT markers for evaluation of genetic diversity and conservation of chickpea (Cicer arietinum L.) landraces genotypes collected from north-west of Iran. Physiol Mol Biol Plant 19:563–574
Pandey MK, Upadhyaya HD, Rathore A, Vadez V, Sheshshayee MS, Sriswathi M, Govil M, Kumar A, Gowda MVC, Sharma S, Hamidou F, Kumar VA, Khera P, Bhat RS, Khan AW, Singh S, Li H, Monyo E, Nadaf HL, Mukri G, Jackson SA, Guo B, Liang X, Varshney RK (2014) Genomewide association studies for 50 agronomic traits in peanut using the ‘Reference Set’ comprising 300 genotypes from 48 countries of the semi-arid tropics of the world. PLoS ONE 9(8):e105228
Perrier X, Flori A, Bonnot F (2003) Data analysis methods. In: Hamon P, Seguin M, Perrier X, Glaszmann JC (eds) Genetic diversity of cultivated tropical plants. Science Publishers, Enfield, pp 43–76
Perrier X, Jacquemoud-Collet JP (2006) DARwin software. https://darwin.cirad.fr/darwin
Pouresmael M, Kanouni H, Hajihasani M, Astraki H, Mirakhorli A, Nasrollahi M, Mozaffari J (2018) Stability of chickpea (Cicer arietinum L.) landraces in national plant gene bank of Iran for drylands. J Agric Sci Technol 20:387–400
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Core Team (2014) R: a language and environment for statistical computing. R Core Team, Vienna
Roorkiwal M, Rathore A, Das RR, Singh MK, Jain A, Srinivasan S, Gaur PM, Chellapilla B, Tripathi S, Li Y, Hickey JM, Lorenz A, Sutton T, Crossa J, Jannink J-L, Varshney RK (2016) Genome-enabled prediction models for yield related traits in Chickpea. Front Plant Sci 7:1666
Roorkiwal M, von Wettberg EJ, Upadhyaya HD, Warschefsky EJ, Rathore A, Varshney RV (2014) Exploring germplasm diversity to understand the domestication process in Cicer spp. using SNP and DArT markers. PLoS One 9:e102016
Saeed A, Darvishzadeh R (2017) Association analysis of biotic and abiotic stresses resistance in chickpea (Cicer spp.) using AFLP markers. Biotechnol Biotechnol Equip 31:698–708
Sagawa CHD, Cristofani-Yaly M, Novelli VM, Bastianel M, Machado MA (2018) Assessing genetic diversity of Citrus by DArT_seq™ genotyping. Plant Biosyst 152:593–598
Saxena MS, Bajaj D, Kujur A, Das S, Badoni S, Kumar V, Singh M, Bansal KC, Tyagi AK, Parida SK (2014) Natural allelic diversity, genetic structure and linkage disequilibrium pattern in wild chickpea. PLoS ONE 9:e107484
Seyedimoradi H, Talebi R, Kanouni H, Naji AM, Karami E (2019) Agro-morphological description, genetic diversity and population structure of chickpea using genomic-SSR and ESR-SSR molecular markers. J Plant Biochem Biotechnol 28:483–495
Sharma S, Upadhyaya HD, Varshney RK, Gowda CLL (2013) Pre-breeding for diversification of primary gene pool and genetic enhancement of grain legumes. Front Plant Sci 4:309
Singh R, Singhal V, Randhawa GJ (2008) Molecular analysis of chickpea (Cicer arietinum L.) cultivars using AFLP and STMS markers. J Plant Biochem Biotech 17:167–171
Talebi R, Fayaz F, Jelodar NA (2007) Correlation and path coefficient analysis of yield and yield components of chickpea (Cicer arietinum L.) under dry land condition in the west of Iran. Asian J Plant Sci 6:1151–1154
Talebi R, Fayaz R, Mardi M, Pirsyedi SM, Naji AM (2008a) Genetic relationships among chickpea (Cicer arietinum) elite lines based on RAPD and agronomic markers. Int J Agr Biol 8:1560–8530
Talebi R, Naji AM, Fayaz F (2008b) Geographical patterns of genetic diversity in cultivated chickpea (Cicer arietinum L.) characterized by amplified fragment length polymorphism. Plant Soil Environ 54:447–452
Thudi M, Khan AW, Kumar V, Gaur PM, Katta AVSK, Garg V, Roorkiwal M, Samineni S, Varshney RK (2016) Whole genome resequencing reveals genome wide variations among parental lines of mapping populations in chickpea (Cicer arietinum). BMC Plant Biol 16:10
Turner NC (1981) Techniques and experimental approaches for the measurement of plant water status. Plant Soil 58:339–366
Upadhyaya HD, Dwivedi SL, Baum M, Varshney RK, Udupa SM, Gowda Cholenahalli LL, Hoisington D, Singh S (2008) Genetic structure, diversity, and allelic richness in composite collection and reference set in chickpea (Cicer arietinum L.). BMC Plant Biol 8:106
Upadhyaya HD, Furman BJ, Dwivedi SL, Udupa SM, Gowda CLL, Baum M, Crouch JH, Buhariwalla HK, Singh S (2006) Development of a composite collection for mining germplasm possessing allelic variation for beneficial traits in chickpea. Plant Genet Resour 4:13–19
Upadhyaya HD, Thudi M, Dronavalli N, Gujaria N, Singh S, Sharma S, Varshney RK (2011) Genomic tools and germplasm diversity for chickpea improvement. Plant Genet Resour 9:45
Van Raden PM (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91:414–4423
Varshney RK, Pandey MK, Bohra A, Singh VK, Thudi M, Saxena RK (2019) Toward the sequence-based breeding in legumes in the post-genome sequencing era. Theor Appl Genet 132:797–816
Varshney RK, Song C, Saxena RK, Azam S, Yu S, Sharpe AG et al (2013) Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat Biotechnol 31:240–246
Yesmin N, Elias SM, Rahman S, Haque T, Mahbub Hasan AKM, Seraj ZI (2014) Unique genotypic differences discovered among indigenous Bangladeshi rice landrace. Int J Genom 2014:210328–210339
Acknowledgements
This study was supported by Azad University, Sanandaj Branch, Iran. We are grateful to Kurdistan Agricultural and Natural Resources and Education Center for providing chickpea genotypes used in this study.
Author information
Authors and Affiliations
Contributions
HS carried out the experiment and prepared the draft of manuscript. RT and AMN conceived and design the experiment, wrote manuscript and analyzed the data. HK prepared the seed material, and EK contributed to data analysis.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Seyedimoradi, H., Talebi, R., Kanouni, H. et al. Genetic diversity and population structure analysis of chickpea (Cicer arietinum L.) advanced breeding lines using whole-genome DArTseq-generated SilicoDArT markers. Braz. J. Bot 43, 541–549 (2020). https://doi.org/10.1007/s40415-020-00634-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40415-020-00634-3