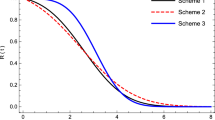
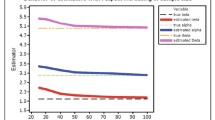
In this article, a constant-stress partially accelerated life test model of high reliability products and materials with type-II censored data from the linear failure rate distribution is considered. Two different methods of estimation likelihood and Bayesian are used to estimate the unknown parameters of the model. The maximum likelihood estimates of the model parameters are obtained using the Newton–Raphson technique. The posterior means and posterior variances are derived under the squared error loss function using Lindley’s approximation procedure. The advantages of this approximation are discussed. Monte Carlo simulations are made for comparing and evaluating the performance of the proposed methods of estimation.


Similar content being viewed by others
References
E. A. Elsayed and A. C. K. Chen, “Recent research and current issues in accelerated testing”, in: Proc. of the 1998 IEEE Int. Conf. on Systems, Man, and Cybernetics (Oct. 11–14, 1998, San Diego, CA, USA), (1998), pp. 4704–4709.
W. Q. Meeker and L. A. Escobar, Statistical Methods for Reliability Data, John Wiley & Sons, New York (1998).
W. B. Nelson, Accelerated Testing: Statistical Models, Test Plans, and Data Analysis, John Wiley & Sons, New York (1990).
Q. Fu and Y. Shi, “Constant-partially accelerated life tests for Marshall–Olkin exponential series system with dependent masked data,” Commun. Stat. Simul. C., 46, No. 10, 7879–7893 (2017).
A. A. Ismail and A. A. Al-Babtain, “Planning failure-censored constant-stress partially accelerated life test,” J. Syst. Eng. Electron., 26, No. 3, 644–650 (2015).
Ali A. Ismail, “Estimating the generalized exponential distribution parameters and the acceleration factor under constant stress partially accelerated life testing with type-II censoring,” Strength Mater., 45, No. 6, 693–702 (2013).
Ali A. Ismail, “On designing constant-stress partially accelerated life tests under time-censoring,” Strength Mater., 46, No. 1, 132–139 (2014).
Ali A. Ismail, “Bayesian estimation under constant-stress partially accelerated life test for Pareto distribution with type-I censoring,” Strength Mater., 47, No. 4, 633–641 (2015).
Ali A. Ismail and A. Al Tamimi, “Optimum constant-stress partially accelerated life test plans using type-I censored data from the inverse Weibull distribution,” Strength Mater., 49, No. 6, 847–855 (2017).
Ali A. Ismail, “Parameter estimation under failure censored constant-stress life testing model using MCMC approach,” Int. J. Ind. Eng.-Theory, 25, No. 3, 402–415 (2018).
Ali A. Ismail, “Planning step-stress life tests for the generalized Rayleigh distribution under progressive type-II censoring with binomial removals,” Strength Mater., 49, No. 2, 292–306 (2017).
Ali A. Ismail and K. Al-Habardi, “On designing time-censored step-stress life test for the Burr type-XII distribution,” Strength Mater., 49, No. 5, 699–709 (2017).
Ali A. Ismail and K. Al-Habardi, “Optimum plans of step-stress life tests using failure-censored data form Burr type-XII distribution,” Strength Mater., 50, No. 4, 674–685 (2018).
J. Jarrahi and H. A. Noughabi, “Testing exponentiality using different entropy estimates based on Type II censored data: A Monte Carlo power comparison,” Int. J. Ind. Eng.-Theory, 24, No. 5, 556–571 (2017).
S. Broadbent, “Simple mortality rates,” J. R. Stat. Soc. C-Appl., 7, 86–95 (1958).
P. O. Carbone, L. E. Kellerhouse, and E. A. Gehan, “Plasmacytic myeloma: a study of the relationship of survival to various clinical manifestations and anomalous protein type in 112 patients,” Am. J. Med., 42, 937–948 (1967).
D. Kodlin, “A new response time distribution,” Biometrics, 2, 227–239 (1967).
D. V. Lindley, “Approximate Bayesian methods,” Trabajos de Estadistica, 31, 223– 245 (1980).
J. Green, “Discussant on D. V. Lindley’s (1980) paper on approximate Bayesian methods,” Trabajos de Estadistica, 31, 241–243 (1980).
S. K. Sinha, Reliability and Life Testing, Wiley Eastern/Halstead, New York (1986).
Author information
Authors and Affiliations
Corresponding author
Additional information
Translated from Problemy Prochnosti, No. 1, pp. 148 – 159, March – April, 2020.
Appendix
Appendix
The derivation of posterior means and posterior variances: the used model contains three parameters. That is, m = 3. Let the subscripts 1, 2, and 3 refer to β, α, and θ, respectively. It is hard to obtain the posterior moments analytically. Therefore, using Lindley’s expansion, the posterior mean (i.e., Bayesian estimator under squared-error loss function) and the posterior variance of β are given, respectively, in the form:
and
Similarly, for the shape parameter α, the posterior mean and the posterior variance are given by
Applying the same technique, the posterior mean and posterior variance of the scale parameter θ take the following form:
and
where
for i, j = 1, 2, 3, σij are the elements of the inverse of the asymptotic Fisher-information matrix of the ML estimators of β, θ, and α in the case of type-II censored data, while i, j = 1, 2, 3, \( {L}_{ijk}^{(3)} \) are the third derivatives of the natural logarithm of the likelihood function in type-II censoring.
To compute the posterior means and the posterior variances of β, θ, and α derived before, both second and third derivatives of the natural logarithm of the likelihood function given in (2) must be obtained.
The second derivatives can be derived via the following equations:
For the third derivatives, they are given as follows:
Rights and permissions
About this article
Cite this article
Ismail, A.A., Al-Harbi, M.M. On the Bayesian Analysis of Constant-Stress Life Test Model Under Type-Ii Censoring. Strength Mater 52, 307–316 (2020). https://doi.org/10.1007/s11223-020-00179-6
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11223-020-00179-6