Abstract
A freshwater alkaliphilic strain of Pseudomonas aeruginosa, grown on waste frying oil-basal medium, produced a surface-active metabolite identified as glycolipopeptide. Bioprocess conditions namely temperature, pH, agitation and duration were comparatively modeled using statistical and artificial neural network (ANN) methods to predict and optimize product yield using the matrix of a central composite rotatable design (CCRD). Response surface methodology (RSM) was the statistical approach while a feed-forward neural network, trained with Levenberg–Marquardt back-propagation algorithm, was the neural network method. Glycolipopeptide model was predicted by a significant (P < 0.001, R2 of 0.9923) quadratic function of the RSM with a mean squared error (MSE) of 3.6661. The neural network model, on the other hand, returned an R2 value of 0.9964 with an MSE of 1.7844. From all error metrics considered, ANN glycolipopeptide model significantly (P < 0.01) outperformed RSM counterpart in predictive modeling capability. Optimization of factor levels for maximum glycolipopeptide concentration produced bioprocess conditions of 32 °C for temperature, 7.6 for pH, agitation speed of 130 rpm and a fermentation time of 66 h, at a combined desirability function of 0.872. The glycosylated lipid-tailed peptide demonstrated significant anti-bacterial activity (MIC = 8.125 µg/mL) against Proteus vulgaris, dose-dependent anti-biofilm activities against Escherichia coli (83%) and Candida dubliniensis (90%) in 24 h and an equally dose-dependent cytotoxic activity against human breast (MCF-7: IC50 = 65.12 µg/mL) and cervical (HeLa: IC50 = 16.44 µg/mL) cancer cell lines. The glycolipopeptide compound is recommended for further studies and trials for application in human cancer therapy.
Similar content being viewed by others
Introduction
Biological amphiphiles with surface and emulsifying activities are called biosurfactants (Ghasemi et al. 2018). They are produced by all life forms including plants (saponins), animals (bile salts) and microorganisms of diverse bacterial, yeast and mold genera, and species (Chen et al. 2010; Bezerra et al. 2018; Kim et al. 2018; Ali et al. 2019). Microbially-derived surfactants are, by far, the most explored, exploited and documented of all three biological sources. The tremendous attention received by them in recent times could be attributed to their huge industrial and environmental applications, which makes them one of the world’s most sought-after bioprocess chemicals besides antibiotics and biocatalysts.
Ability of biosurfactants to solubilize and emulsify hydrophobic organic compounds through surface and interfacial tension reduction, forms the basis for their applications in enhanced recovery of oil as well as in enhanced bioremediation of crude oil-impacted ecosystems (Bezza et al. 2015; Almeida et al. 2016; Bezza and Chirwa 2017; Ali et al. 2019). Their foaming or detergency property makes them good candidates for production of detergents and personal care products (Terziyski et al. 2014; Vecino et al. 2018) while their chelating property has found applications in bioremediation of toxic metal-impacted environments and in leaching of metals from their ores (Alsaqer et al. 2018; da Rocha Junior et al. 2019). Biosurfactants demonstrate commendable biological activities including antimicrobial, anti-adhesive, anti-biofilm and anti-cancer activities which are currently being exploited in medicine, food, cosmetic and pharmaceutical industries, as well as in plant-pathogen control in agriculture (Sivapathasekaran et al. 2010a, b; Wu et al. 2017; Vecino et al. 2018; Satpute et al. 2018a, b; Jimoh and Lin 2019; Bertrand and Munoz-Garay 2019; Javee et al. 2020; Rani et al. 2020).
Microbial surfactants occur in diverse chemical types such as glycolipids, lipopeptides, phospho- and flavo-lipids, and the polymeric and particulate types. Production of a particular biosurfactant type is not the exclusive preserve of any microbial genus or species. Among the bacterial genera, Pseudomonas and Bacillus biosurfactants have been most studied and documented (Perfumo et al. 2017). Rhamnolipid; a type of glycolipid biosurfactant; has, until recently, been reported almost exclusively in Pseudomonas aeruginosa strains. However, Hoskova et al. (2015) have characterized rhamnolipids from Acinetobacter calcoaceticus and Enterobacter asburiae. Biosurfactant types other than rhamnolipid, including protein PA (Hisatsuka et al. 1972), glycolipopeptide (Koronelli et al. 1983; Ilori and Amund, 2001; Ekpenyong et al. 2016) and lipopeptide (Thavasi et al. 2011a, b) have also been reported in a few special strains of Pseudomonas aeruginosa. Other species like Pseudomonas fluorescens and P. putida have been reported to produce glycolipopeptide (Desai et al. 1988), and lipopeptide and rhamnolipid (Kuiper et al. 2004; Martinez-Toledo and Rodriguez-Vazquez 2013), respectively. Bacillus species are most commonly reported to produce lipopeptides. However, there have been reports of some species that produce glycolipid and glycolipopeptide biosurfactants as well (Thavasi et al. 2008; Mabrouk et al. 2014).
Glycolipopeptides are a form of lipid-tailed polymeric biological amphiphile produced by a few bacterial genera and/or species and have been categorized as bio-emulsifiers rather than biosurfactants on the basis of their high molecular weights (Gutierrez and Banat 2015). They are glycosylated lipopeptides or lipid-tailed glycoproteins commonly noted for producing stable water-in-oil or oil-in-water emulsions with hydrophobic compounds (Satpute et al. 2016). Reported microbial genera associated with their production include Pseudomonas, Bacillus, Corynebacterium and Lactobacillus. Glycolipopeptides from Lactobacillus species are mostly cell-associated but those from other genera are not (Vecino et al. 2018). There has been a heightened interest in glycolipopeptide biosurfactants arising from their demonstration of biological activities including antimicrobial, anti-biofilm and anticancer activities (Karlapudi et al. 2020; Javee et al. 2020). A synthetic form of this compound is currently under investigation as potential vaccine base for B- and T-cell immunotherapy for cancers (Renaudet et al. 2010).
Global availability of biosurfactants for their much-needed applications has been limited by the same bioprocess economics frequently encountered in typical microbial metabolite production processes. The specific variables required to develop a successful biosurfactant fermentative production process include strain selection and improvement (Hu et al. 2019; El-Housseiny et al. 2019), medium and process development through optimization (Rodrigues et al. 2019) and, identification and improvement of catalytic rates towards scale-up processes to maximize volumetric productivity (Truppo 2017; Wachtmeister and Rother 2017).
Design of experiments (DoE) has facilitated the development of fermentation media for biosurfactant production where major and trace nutrients composition, as well as environmental and/or operational conditions have been studied using different optimization protocols. A few decades ago, the traditional one-factor-at-a-time (OFAT) technique dominated literature and still applies today in the early stages of bioprocess optimization (Nicolo et al. 2017). This method, with its inherent limitations of tedium and inability to account for interaction effects between and among independent variables, has been largely replaced with more robust techniques like response surface methodology (RSM) and artificial neural network (ANN) (Bertrand et al. 2018).
Response surface methodology (RSM) refers to a collection of statistical and mathematical techniques for constructing approximation models based on physical experimentation or computer simulation. The primary objective of RSM is to optimize the response whose magnitude and direction is controlled by predictor variables. With RSM, optimization of medium and bioprocess conditions for microbial metabolite fermentations have been improved (Rodrigues et al. 2006; Ebadipour et al. 2016; Chouaibi et al. 2019). Artificial neural network (ANN) approaches, on the other hand, are intelligence-based modeling techniques that rely on biological information processing to build models. They are a computer-based system that employs a number of neurons to simulate the learning pattern of the human brain. The neurons are grouped into one or more hidden layers that are connected using synapses called weights. ANN approaches are particularly suited to stochastic situations such as production since they typically detect the overwhelming non-linear relationships to facilitate a better modeling of the process (Sivapathasekaran et al. 2010a, b; Santos et al. 2017). Very often, these two optimization methods are compared for superiority and majority of the reports tilt in favour of ANN methods without consideration of the objective of research and tact of the experimenter (Pal et al. 2009; Patel and Brahmbhatt 2016; Sampaio et al. 2017; Karri et al. 2018).
In the present study, performances of RSM and ANN as techniques to build predictive models and optimize fermentation conditions that enhance production of bioactive glycolipopeptide surface-active agent by Pseudomonas aeruginosa strain IKW1 are reported. Fermentation conditions including temperature, pH, agitation speed and fermentation time were the input variables while biomass and glycolipopeptide concentrations were the responses. Model performance was evaluated with error metrics other than coefficient of determination. Results are expected to guide the development of appropriate operational conditions for fermentative biosurfactant production and facilitate the determination of optimum kinetic and economic parameters in future researches towards scale-up of the bioprocess.
Materials and Methods
Inoculum Preparation
The bacterium, Pseudomonas aeruginosa strain IKW1, earlier isolated by Ekpenyong et al. (2016) and deposited with the University of Calabar Collection of Microorganisms (Reg. No: WDCM 652), was retrieved from the culture collection and reactivated for the study using the protocols described in Ekpenyong et al. (2017a).
Medium Development and Preparation
Fermentation medium was formulated as per the optimum major medium factor levels reported in Ekpenyong et al. (2017a) and trace element settings in Ekpenyong et al. (2017b). The medium contained 5% (v/v) carbon source of waste frying oil. The waste oil composed of 2.18% w/w steric acid, 16.14% w/w volatile fractions; 22.51% w/w oleic acid; 50.63% w/w linoleic acid; 6.13% w/w palmitic acid; a saponification value of 73 and density of 286.7 kg/m3 at 30 °C. Phosphates and buffering capacity were supplied as 4.5 g/L Na2HPO4/KH2PO4 (2:1); 1.19 g/L filter-sterilized urea (Merck) served as nitrogen source; nutritive salts were supplied as 0.2 g/L MgSO4.7H2O, 0.5 g/L NaCl, 0.5 g/L CaCl2, 0.5 g/L KCl. One milliliter of a solution of trace metals comprising 1.2500 mg nickel/L, 0.1250 mg zinc/L, 0.1250 mg iron/L, 0.0104 mg boron/L and 0.0250 mg copper/L was used to fortify the production medium (Ekpenyong et al. 2017b). The medium, without nitrogen source, was dispensed into 250 mL Erlenmeyer flasks (20% v/v) and sterilized by moist heating. The nitrogen source, urea (1.19 g/L) was sterilized by filtration and incorporated into each flask after cooling to 40 °C. The pH was adjusted as per experimental design using sodium-phosphate buffer (pH 5 to 6.5) and Tris–HCl (pH 7.0 to 8.4) (Edet et al. 2018). Thereafter, 10% (v/v) of triple-washed (phosphate buffered saline, pH 7) suspension (108 cfu/mL) of an overnight LB broth culture of Pseudomonas aeruginosa strain IKW1 (UCCM 0002) was added to each flask. Each flask was plugged with sterile cotton wool and incubated on a rotary shaker as per experimental design for temperature, agitation speed and duration (Table 1).
Experimental Design and Incubation Protocol
A 24 full-factorial central composite rotatable design (CCRD) was adopted as experimental design using Design Expert 9. Table 1 presents the actual and coded levels for the design matrix of the four input variables for the CCRD. The factors were earlier identified by Ekpenyong et al. (2016) as significant bioprocess operational conditions that influenced glycolipopeptide biosynthesis by the bacterium. The coded levels were determined as follows; X1 = (temperature-30)/2; X2 = (pH-7.0)/0.6; X3 = (Agitation speed-150)/20 and X4 = (Fermentation time-72)/6. The actual levels, on the other hand, were calculated using the equation below (Myers and Montgomery 2002):
The design matrix required 30 experimental runs. Incubation, at test bioprocess conditions, was adjusted according to experimental design (Table 1). Each experimental run was prepared in triplicates. Concentrations (g/L) of biomass (Y1) and glycolipopeptide (Y2) served as study outcomes.
Estimation of Concentrations of Biomass and Glycolipopeptide
Aliquots, measuring 10 mL from each experimental run, were withdrawn at intervals according to design. Biomass and surface-active compound were harvested by centrifugation of flask contents at 8000×g for 15 min. Membrane filtration was adopted to sterilize the supernatant and glycolipopeptide was recovered from resulting sterile filtrate by acid precipitation at pH 2.0 using 6 N HCl, solvent extraction, dialysis and vacuum drying. Quantification of biomass followed the dry weight technique (Ekpenyong et al. 2016).
Modeling and Optimization Protocols
Response Surface Methodology (RSM)
Experimentally-generated data were analyzed by multiple regression analysis. Regression models for biomass and glycolipopeptide responses of a surface methodology were built by the least squares method at 5% significance level using Design Expert 9. Data was fitted using second-order polynomial function. Contour, surface and cube plotting, as well as determination of the optimal levels of factors using the desirability function, were performed by the statistical software used for the design of experiment (DoE).
The general quadratic model for the response variables took the form below;
where β0 denotes a constant coefficient, k the coefficient of the linear effect of the kth factor, βii the quadratic effect of the ith factor and βij the effect of the interaction between the ith and jth factors, where x1, x2, … xk are the independent variables and ε the error arising from the computation of the response variable Y.
Artificial Neural Network (ANN) Modeling
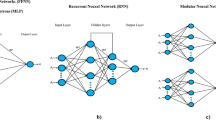
A two-layer feed-forward neural network with sigmoid hidden neurons trained with Levenberg–Marquardt back-propagation algorithm was employed to model the response data generated from the CCRD against the predictor variables. MATLAB R2014a software (Mathworks Inc, Natick, USA) was used to create the network. The network topology was 4-9-1 corresponding respectively, to the number of neurons in the input layer, hidden layer and output layer, to establish the influence of the four fermentation conditions of temperature, pH, agitation and fermentation time on biomass or glycolipopeptide concentrations. The variables in the input and output layers were scaled as [− 1, 1].
A total of 30 samples were presented to the network, 70% of which were used to train the network, 15% to validate its performance and another 15% to test it. Performance of the network, defined as the closeness of the model to real life systems, was evaluated with the values of coefficient of determination (R2) and mean squared error (MSE). To facilitate the comparison between the two models, root mean squared error (RMSE), mean absolute error (MAE), mean absolute percentage error (MAPE), mean percentage error (MPE) and Pearson’s Chi-squared measure (χ2) of both models were determined using Eqs. 3–11 below.
where n is the number of samples, y the actual observed value, \({\hat{{{y}}}}\) the predicted value and \({\bar{{{y}}}}\) the mean of the actual value, y.
Validation Experiments
Experiments were set up to validate the optimum operational conditions of temperature, pH, agitation and fermentation time suggested by the response optimizer in Design Expert 9. Fermentation medium composition, incubation and outcome determinations were as previously described in their respective sections in this study.
Determination of Biological Activities of Glycolipopeptide
Evaluation of Antimicrobial Activity of Glycolipopeptide
Test microbial strains included Streptococcus mutans UCCM 0047 and Bacillus cereus UCCM 0007 (Gram-positives), Escherichia coli ATCC 10536 and Proteus vulgaris ATCC 27973 (Gram-negatives), Fusarium oxysporum UCCM 0077 and Aspergillus fumigatus UCCM 0052 (molds) and Candida albicans ATCC 10231 and Cryptococcus neoformans CAB1034 (yeasts).
Bacterial suspensions of 106 cfu/mL and yeast suspension of 103 cfu/mL were prepared by the spectrophotometric method. Spore suspensions of molds were prepared in Czapek–Dox agar (CDA) in Roux bottles and incubated at 35 °C for 14 days. Spores were dislodged from mycelia by gentle tube inversions after adding 20 mL cold distilled water mixed with glass beads. The suspension was washed twice with cold water by centrifugation at 5000 rpm for 5 min. Spores were suspended in 2 mL of cold water to form the concentrated stock suspension. Test concentration (103 cfu/mL) of each mold was prepared from the concentrated stock solution by the spectrophotometric method.
For determination of minimum inhibitory concentration (MIC) of the compound, the broth microdilution technique of Clinical and Laboratory Standards Institute (CLSI) was adopted. Muller Hinton broth (Himeda, India) was prepared according to document M7-A6 for bacterial MICs and M27-A3 for fungal MICs (CLSI 2019) Glycolipopeptide concentration was prepared in consideration of its critical micelle concentration (CMC) of 20.80 mg/L given in Ekpenyong et al. (2016). Test compound concentration of 2.08 mg/mL (2080 µg/mL) was prepared in distilled water and diluted twofold in Muller-Hinton broth to obtain 1040 µg/mL stock solution. Working concentrations of the test glycosylated lipopeptide ranging from 1.015625 to 520 µg/mL were prepared by twofold serial dilution using Muller-Hinton broth as diluent. A polystyrene panel of 96-wells was used with two wells serving as positive (broth and inoculum) and negative (broth without inoculum) controls. Chloramphenicol and amphotericin B (Sigma Aldrich, USA) served as positive bacterial and fungal controls respectively. For tests involving fungi, glycolipopeptide stock solution was prepared in RPMI 1640 (Sigma Aldrich). An aliquot of 0.1 mL of each of bacterial, yeast and mold spores were inoculated into each well to give 5 × 104 cfu/well, 2.5 × 102 cfu/well and 2.5 × 102 spore forming units (sfu) per well respectively. All arrangements were made in triplicates and plates incubated at 37 °C for 24 h, 48 h and 60 h for bacteria, yeasts and mold determinations respectively. The minimum inhibitory concentration (MIC) was defined as the lowest concentration of the glycolipopeptide that could completely inhibit visible growth of test organism under study conditions.
Evaluation of Anti-biofilm Activity of Glycolipopeptide Using the Crystal Violet Quantification Method
Ability of the glycolipopeptide to disrupt previously formed biofilms of two bacteria and two yeast species were tested. Selected biofilm-forming organisms included Enterococcus aerogenes UCCM 0089, Escherichia coli ATCC 10536, Candida albicans ATCC 10231 and Candida dubliniensis UCCM 0058. A 48-h old Sabouraud dextrose agar culture of Candida species was used to inoculate 50 mL of yeast nitrogen base (YNB) medium supplemented with 1% glucose in 250-mL Erlenmeyer flasks and incubated for 24 h at 37 °C in an orbital water-bath shaker at 60 rpm. Cells were harvested by centrifugation at 6,000 rpm and washed twice with 0.15 M magnesium-free phosphate-buffered saline (PBS; pH 7.2). Cells were re-suspended in 10 mL of PBS and standardized to a concentration equivalent to 0.5 McFarland standards and used immediately. Brain heart infusion (BHI) broth (180 µL) containing 1% glucose was mixed with 20 µL of standardized bacterial culture while 180 µL of RPMI 1640 was also mixed with 20 µL of standardized yeast culture. Preparations, made in five replicates, were incubated in 96-well polystyrene plates for 24 h at 37 °C for bacteria and 30 °C for yeasts. Plates were washed with PBS after incubation to remove unattached cells. Thereafter, plates were treated with three concentrations of glycolipopeptide; 45, 90 and 180 µL/mL and incubated as previously described. Untreated wells that received only PBS served as positive control. All unattached cells were removed by washing five times with PBS and by use of pipette. Biofilms were defined as cells that attached to the bottom of wells. The biofilms were fixed with 300 µL of 99% methanol and stained with 1% crystal violet solution for 5 min. Excess stain was removed by washing with running tap water. Plates were allowed to dry in air after which they were treated with 200 µL of 95% ethanol to dissolve the dye. The absorbances of solutions obtained were read off a 340 Microplate Reader (Thermo Scientific, USA) at 450 nm wavelength. Per cent biofilm disruption was calculated by comparing the absorbance of treated biofilm with the un-treated control (Morais et al. 2017).
Evaluation of Anti-cancer Activity of Glycolipopeptide
The cell proliferation method based on the MTT assay protocol of Sivapathasekaran et al. (2010a, b) was adopted. The glycolipopeptide was tested against human breast cancer (MCF-7), leukemia (HL-60) and cervical epithelial carcinoma (HeLa) cell lines (ATCC) at concentrations ranging from 5.20 to 665.6 µg/mL. The concentrations were twofold dilutions of concentrated dialyzed fractions containing 665.6 µg/mL of compound chosen as a multiple of the critical micelle concentration of the glycolipopeptide reported elsewhere (Ekpenyong et al. 2016). Cancer cell lines were grown to initial cell density of 105 cells/well in a 96-well microtiter plate. All preparations were made in triplicates. Selective inhibitory effect of the peptide compound was tested against a non-tumor human (HEK 283 T) embryonic cell line. Wells without glycolipopeptide served as positive controls with 100% cell viability. The highest concentration of glycolipopeptide that recorded no viable cell served as negative control with 0% viability. The formed purple-coloured product was dissolved in dimethyl sulfoxide and its absorbance read off at 570 nm wavelength. Non-linear regression analysis of data, graphing and determination of median inhibitory concentration (IC50) of glycolipopeptide against all cancer cell lines was conducted using GraphPad Prism 8.
Determination of Surface Activities of Glycolipopeptide
Surface Tension Determination
Surface tension reduction potential of the glycolipopeptide which indicates surfactant effectiveness and the critical micelle concentration (cmc) which indicates its efficiency were determined as described in Ekpenyong et al. (2016).
Determination of Oil Displacement Activity
The oil displacement activity of the glycolipopeptide was determined by the oil spreading method of Morikawa and co-workers as described in Ekpenyong et al. (2016).
Determination of Emulsifying Activity
The emulsifying activity of the glycolipopeptide was measured by mixing equal volumes of each of palm oil and kerosene with equal volume of sterile glycolipopeptide broth and homogenizing for 5 min (Ekpenyong et al. 2016). The resulting emulsion was allowed to stand in the ambient conditions of the laboratory for 24 h and the percentage ratio of the height of emulsion to the total height of liquid in the tube was scored as emulsification index 24 (E24).
Results and Discussion
Study Design Observations
Table 2 is a presentation of the experimental observations for response variables; biomass, Y1 and glycolipopeptide concentrations, Y2 obtained from the central composite rotatable design (CCRD). Highest glycolipopeptide concentration of 117.2 g/L was obtained in run 10 (randomized as 3) while the least concentration of 50.87 g/L occurred in run 18 (randomized as 24). Highest glycolipopeptide concentration corresponded to bioprocess conditions of temperature-28 °C, pH-7.6, agitation speed-130 rpm and duration 66 h. Lowest glycolipoepeptide concentration was obtained under conditions set at tempearture-30 °C, pH-7, agitation-150 rpm and duration-84 h. When these conditions were compared with previously reported conditions in Ekpenyong et al. (2017a, b), it was observed that the first three bioprocess conditions of temperature, pH and agitation had similar settings, however, duration of fermentation, when extended from 72 to 84 h reduced glycolipopeptide concentration from 84.44 g/L (Ekpenyong et al. 2017b) to 50.87 g/L. It was observed, for this run (data not shown), that the fermentation broth became viscous by the 80th hour suggesting possible release of exopolysaacharide by the bacterium. Production of exopolysaccharides by Pseudomonas aeruginosa strains have been reported by Franklin et al. (2011) as a measure to protect cells against environmental stress, in this case nutrient deprivation, by self-encapsulation within the matrix material.
RSM Models
Analysis of Variance of Models
The ANOVA for biomass model is presented in Table 3 and revealed an adjusted R2 value of 0.9887 suggesting that 98.87% of the variations about the data could be explained by the model. The table also showed that four predictor interactive terms, namely, X1X4, X2X3, X2X4, X3X4, did not contribute significantly to the model. This indicated that the model had redundant predictor terms whose addition or removal did not significantly improve the model. The RSM model equation for biomass concentration, after elimination of the non-significant predictor terms, is therefore presented as Eq. 10 below;
The equation however, showed that the linear term of pH (X2) made significant positive (enhancement) effect on biomass concentration, Y1 whereas its quadratic term significantly reduced biomass concentration. All the quadratic terms had significant effect on biomass concentration, however, only the quadratic effect of agitation enhanced biomass accumulation.
The ANOVA summary describing glycolipopeptide regression model predictor variables is presented as Table 4. The model, after eliminating non-significant predictor terms, is given by the equation below;
Once again, the pH (X2) made significant enhancement effect on Y2 (glycolipopeptide concentration). However, if the coefficient of predictor X2 in Eq. 6 is compared with that in Eq. 7, a higher enhancement influence of pH on glycolipopeptide concentration (10.99) than on biomass concentration (2.8) is observed suggesting that the environmental factor had more influence on metabolite synthesis than biomass accumulation. Additionally, whereas the quadratic term of the predictor variable pH reduced biomass concentration, the opposite was the case for glycolipopeptide concentration, Y2. This suggests that the optimal levels of pH are not in the extremes of the experimental region of the glycolipopeptide model but within it.
The linear term of temperature reduced glycolipopeptide concentration while the quadratic term enhanced its production. These results suggest that conditions for cellular growth of microorganisms are different in the vast majority of cases from those required for metabolite biosynthesis. Ekpenyong et al. (2017a) had earlier reported this in their research on major nutritional parameters that influence glycolipopeptide production. In a batch production system such as employed in this study, the only way to go around this problem would be to identify the metabolite of interest and optimize conditions that favour it.
Contour, Surface and Cube Plots of RSM Models
A contour plot is a 2-dimensional representation of the surface with similar responses connected to produce desirable response values and operating conditions. Figure 1a shows a two-way interaction effect between pH and temperature while the levels of agitation and duration of fermentation were held at their mid-points. The figure indicates very marginal curved contours suggesting a ridge-shaped or planar surface of a linear model. The 3D surface plot, on the other hand, displays a surface view in three dimensions to establish desirable response values and operating conditions but provide a clearer concept of the response surface than 2D plots. Figure 1b shows the 3D plot involving the interaction between pH and temperature when the levels of agitation speed and duration of fermentation were held at their mid-points. The 3D plot reveals a rising ridge surface indicating increasing response with increased darkening of colour. This suggests that biomass response increased as pH increased but with decreasing temperature. Maximum biomass was obtained when at high pH level of 0.5.corresponding to an actual value of 7.6 but at low temperature of -0.5 corresponding to an actual value of 28 °C.
Figure 2a, b present the contour and surface plots for glycolipopeptide regression model respectively. Figure 2a shows a 2D plot where the contour lines are spaced far apart from each other suggesting that the glycolipopeptide response values changed quite slowly. Maximum glycolipopeptide was obtained when the pH was high with decreasing temperature. The 3D plot in Fig. 2b for glycolipopeptide regression model reveals a planar surface with slow changes in glycolipopeptide concentration. High pH favoured high glycolipopeptide concentration but at slowly decreasing temperature.
A more robust explanation of interaction effects is presented by cube plots in Fig. 3a, b. Figure 3a indicates that pH and process temperature interacted to affect the biomass response. When pH increased from low level to high level and temperature was low, the increase in biomass concentration was about 2.76 g/L. However, when temperature increased and the pH was low, biomass concentration dropped by 7.38 g/L.
This implies that high temperatures are not salutary to the accumulation of Pseudomonas aeruginosa strain IKW1 biomass when the pH was low as earlier suggested by the contour and surface plots. However, since the model has interaction between pH and temperature, the effects were different when the variables were at their high levels. When temperature increased and pH was high, the drop in biomass concentration was only about 1.76 g/L. But when the pH increased and temperature was high, the drop in biomass increased by about 8.44 g/L. Figure 3a also includes agitation as a third predictor of biomass model which agrees with the results presented in the analysis of variance table for biomass model (Table 3). The cube plot confirmed that increase in the bioprocess speed of agitation caused a negative change in biomass concentration by a factor of 0.92 g/L.
Figure 3b presents the cube plot for glycolipopeptide regression model and reveals that when pH increased to high level and bioprocess temperature was low, glycolipopeptide concentration increased by 18.43 g/L, but when temperature increased from low to high levels and the pH was low, concentration of glycolipopeptide dropped by about 12.96 g/L. That reduction was large and to avoid that, both parameters were held at their high levels. When the temperature increased and pH was high, the reduction in glycolipopeptide concentration was only 6.22 g/L. On the other hand, if the pH increased and temperature was high, the glycolipopeptide concentration increased by 25.17 g/L. Since this research is about improving glycolipopeptide yield, then the incremental raise in pH at high temperature, with low process agitation speed would be most salutary for maximum glycolipopeptide production.
ANN Models for Response Variables
Results of the two-layer feed-forward neural network are presented in Fig. 4. Figure 4a is the topology of the network and depicts the number of neurons deployed in the various layers of the network. Figure 4b shows how the network that trained samples for biomass model performed. While the training samples had a mean squared error (MSE) of 0.005933 and an R2 value of 0.9996, the validation samples had an MSE of 0.27969 and R2 of 0.9892 and the testing samples had an MSE of 0.327546 and an R2 of 0.9662. However, when the overall network was tested, a performance evaluation result for biomass model was R2 = 0.9971; MSE = 0.10516; RMSE = 0.3240. These values were obtained using 9 iterations and validation performance value of 0.2797 reached at epoch 6.
Figure 4c, on the other hand presents the network performance for glycolipopeptide model. The training samples had an MSE of 0.2581 and an R2 of 0.9988; validation samples had an MSE of 7.6320 and an R2 of 0.8881 while the testing samples had an MSE of 2.04238 and an R2 of 0.9997. However, when the overall network was tested for its performance, the glycolipopeptide model had an R2 of 0.9962; MSE = 1.7844; RMSE = 3.184. These values were obtained using 7 iterations with a performance value of 7.632 at epoch 5. These results indicate that the error of prediction by the ANN approach was higher for the glycolipopeptide than for biomass model suggesting the involvement of more predictors for glycolipopeptide concentration.
The predicted values for biomass and glycolipopeptide models obtained with ANN are presented in Table 5. Maximum biomass concentration predicted by the network was 28.03 g/L when fermentation conditions were set at 30 °C-temperature, 8.2-pH, 150 rpm-agitation speed and a fermentation time of 72 h but had its minimum as 15.72 g/L when fermentation conditions were set at 32 °C, pH of 6.4, agitation speed of 170 rpm and a fermentation time of 78 h. On the other hand, maximum concentration of glycolipopeptide predicted by the ANN model was 120.06 g/L when operating conditions were set at 28 °C, pH of 7.6, agitation speed of 130 rpm and a fermentation time of 66 h. However, the model had its minimum of 50.88 g/L when fermentation conditions were set at 30 °C, pH of 7.0, agitation speed of 150 rpm and duration of operation of 84 h. The ANN model confirmed the significance of prolonged fermentation on glycolipopeptide production as counter-productive as the compound of interest has a tendency of being converted to exopolysaccharides for bacterial protection (Franklin et al. 2011).
Comparison Between RSM and ANN Models
Comparison of the two approaches was based predominantly on their predictive capabilities using seven model performance parameters namely R2, MSE, RMSE, MAE, MAPE, MPE and Pearson’s χ2. Consideration for this number of performance metrics became unavoidable seeing that R2 alone as a model goodness-of-fit was not sufficient as observed in the similarity of regression plots in Fig. 5. The results of glycolipopeptide model comparison performed by multiple t-tests using the two-stage linear step-up procedure are presented as Fig. 6. Figure 6a shows negative MPE which underlies underperformance of models. All the error metrics significantly (P = 0.01) contributed to the performance of the model. Briefly, the MSE is a metric which ensures that the model takes into account possible outliers (large residuals between actual and predicted values) of the model but to play down on the significance of outliers, the MAE metric is used. Determination of MSE requires that squared residuals be used for computation (Eq. 4) which further increases the magnitude of outliers and strengthens the discriminating effect of the metric. Since outliers are effects observed naturally, MSE serves model performance evaluation better with its quadratic contribution to total error than MAE which contributes proportionally, where outliers are a problem, otherwise MAE should do. All the evaluation metrics were in favour of ANN as a superior predictive modeling approach. The negative MPE values for both models indicated underperformance of both models. Under-performing models are recommended since they do not give false expectations like over-performing models. Figure 6b is a volcano plot of the error metrics with a superimposed multiple t-test analysis results table. The table arranges the t-ratios in order of descending magnitude and reports the q value for each. The q-value is the p-value which has been adjusted for the proportion of false positives called the false discovery rate (FDR), which usually escape detection by the p-value. The negative logarithm of the q-values gives the most significant error metrics for model performance evaluation as MAE, MSE, Pearson’s Chi-Squared measure and RMSE in that order, and the least significant of all measures as the R2 metric (Fig. 5e). Several researches are in agreement with our findings that the ANN approach is a better modeling approach in terms of prediction capability than RSM (Pilkington et al. 2014; Patel and Brahmbhatt. 2016; Karri et al. 2018). The major advantage that ANN has over RSM as a prediction modeling tool is its overwhelming ability to handle non-linear situations between response and independent variables (Patel and Brahmbhatt 2016). The almost-always limitation of RSM to quadratic models makes ANN a better modeling approach. If the cubic model is suggested for an RSM, then it would be a better modeling and optimization tool than ANN or just as good (Sampaio et al. 2017).
Comparison of ANN and RSM models for glycolipopeptide production using a bar charts of a multiple t-test of model error metrics and b volcano plot of q values against individual error metric difference. The inserted table presents the magnitude of q values, with the encircled ones recommended as more reliable comparison error metrics
Statistical Optimization of Bioprocess Conditions for Maximum Glycolipopeptide Concentration
The result of the statistical optimization of process conditions for maximum glycolipopeptide as goal is presented in Fig. 7a. The figure shows that biomass (Y1) and glycolipopeptide (Y2) concentrations of 25.30 g/L and 107.19 g/L respectively were obtained at (X1, X2, X3, X4) = (1, 1, − 1, − 1), corresponding to actual values of temperature-32 °C, pH-7.6, agitation-130 rpm and fermentation time-66 h, with a production yield, Yp/x of 4.24. The desirability plot is given as Fig. 6b and revealed a desirability function of 0.872. Desirability is an objective function whose value ranges from zero to one. The closer the function is to 1, the more reliable the results of the optimization process. To obtain a single desirability function in a production study such as this, where more than one goal could be involved, the desirable functions of the individual goals are frequently combined into one desirable function.
Validation of Suggested Optimum Fermentation Conditions
Experiments to validate the optimum predictor levels suggested in Sect. 3.2, returned a glycolipopeptide concentration of 108.54 g/L and a biomass concentration of 25.49 g/L which gave a product yield, Yp/x of 4.26. By comparing, especially the glycolipopeptide concentration of 107.19 g/L given by the response optimizer with that obtained in the validation experiment, the difference between them was not up to 1%. Therefore, it was concluded that the optimum level settings for the bioprocess conditions required to maximize glycolipopeptide production by the bacterium were adequate.
Antimicrobial Activities of Glycolipopeptide Compound
The glycolipopeptide exhibited a dose-dependent inhibition of Streptococcus mutans, Escherichia coli, Proteus vulgaris, Candida albicans, Fusarium oxysporum and Aspergillus fumigatus. However, the metabolite did not inhibit Bacillus cereus and Cryptoccocus neoformans at any of the tested concentrations. Table 6 presents the MICs of the peptide compound. Proteus vulgaris was most inhibited by the compound with an MIC of 8.125 µg/mL. The mold and yeast organisms had large MICs of 130 µg/mL, suggesting that larger concentrations of the compound could be required to disrupt fungal functions. Not even the powerful lipopeptide biosurfactant, surfactin could inhibit Cryptococcus neoformans CAB1034 at 260 µg/mL by the disk diffusion assay (Nicolo et al. 2017). It is expected that a fully-purified compound will have a better activity than what is presented in this study. Inhibition of Fusarium oxysporum by the glycolipopeptide could be exploited in plant-pathogen control as the mold is noted for wrinkling of fluted-pumpkin leaves; a foremost edible leaf in the West-African region, especially South-South Nigeria. The MIC of 8.125 µg/mL of the partially-purified compound is informative and points to its potency in the control of Proteus vulgaris associated diseases with a fully-purified product. Reports on microbial production of glycolipopeptides are few (Thavasi et al. 2011a; Karlapudi et al. 2020; Hippolyte et al. 2018), and even fewer address biological activity potentials. Kiran et al. (2009) reported a similar broad spectrum antimicrobial potential of a glycolipoprotein produced from a sponge-associated marine fungus Aspergillus ustus strain MSF3. Both Karlapudi et al. (2020) and Hippolyte et al. (2018) used the agar well diffusion method as against the broth microdilution method adopted in this study and so the MIC results could not be corroborated. However, all four reports have confirmed the antimicrobial activity of their various glycolipopeptides.
Anti-biofilm Activities of the Partially-Purified Fraction of Glycolipopeptide
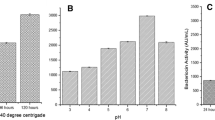
Result of the anti-biofilm activity of the glycolipopeptide metabolite is presented as Fig. 8 and showed a dose-dependent inhibitory activity of the compound. Percent inhibition of biofilm formation was found to be directly proportional to concentration of bioactive compound. However, degrees of bio-film inhibition varied with respect to test microorganism that formed it suggesting that biofilms are chemically and physically diverse as they are biologically complex. A two-way ANOVA revealed that the interaction effects between test compound concentration and bacterial type was significant, F (2.982, 11.93) = 24.68, P < 0.0001, with a Geisser Greenhouse epsilon correction value for assumption of no sphericity of 0.497. Figure 8 clearly shows that Escherichia coli and Candida dubliniensis were the most susceptible biofilms to glycolipopeptide treatment. Concentrations as low as 45 µg/mL could inhibit formation of their respective biofilms by 38.4% and 30.5% within 24 h while 180 µg/mL resulted in 83.7% and 90.6% inhibitions respectively. Karlapudi et al. (2020) reported a biofilm inhibition of 82.5% in methicillin-resistant strain of Staphylococcus aureus at 500 µg/mL of glycolipoprotein compound from Acinetobacter indicus strain M6.
Anti-cancer Activities of the Partially-Purified Fraction of Glycolipopeptide
Certain types of glycoproteins are reported to be overexpressed during cancers and have therefore been targets for vaccine development. Lipid-tailed glycoproteins (glyco-lipo-peptides) have been developed as a higher ordered structure of those vaccines with higher specificity towards cancer cells and more protection for healthy cells (McDonald et al. 2015). Discovery of naturally-occurring anti-cancer glycolipopeptides from microorganisms could be a major breakthrough in cancer research. Glycolipopeptides’ cytotoxic activity is reported to proceed through activation of a complement-dependent lysis of MCF-7 breast cancer cells. In the present study, the partially-purified glycolipopeptide compound exhibited a dose-dependent inhibition of cancer cell proliferation. Details of the result are presented as Fig. 9. The sigmoidal non-linear regression with least squares fit gave half maximal inhibitory concentration (IC50) of 6.025 ± 0.053 (65.12 µg glycolipopeptide/mL) to MCF-7 cell line (Fig. 9a), 4.039 ± 0.052 (16.44 µg/mL) to HeLa cell line (Fig. 9b) and 8.439 ± 1.811 (347.05 µg/mL) to HL-60 cell line (Fig. 9c). The plot for HL-60 cell line (Fig. 9c) was not sigmoidal and the model showed significant lack-of-fit (F = 3.571, P = 0.0203) suggesting inadequacy to explain data variations about the region of experimentation. The glycolipopeptide is therefore not suitable for treatment of leukemia cell lines (at least not in its partially-purified state). The non-significant lack-of-fit of the non-linear regression sigmoidal models for cytotoxic effect of glycolipopeptide on MCF-7 and HeLa cell lines (F = 1.525, P = 0.2316, R2 = 0.9947; F = 2.313, P = 0.1204, R2 = 0.9955) suggested model adequacy for explanation of variations about the data. The peptide compound demonstrated selective inhibition by its very large IC50 (> 665.6 µg/mL) on the per cent viability of normal non-tumor human embryonic cell line (HEK 283T). Rhamnolipid biosurfactants have been reported to demonstrate lower IC50 values of 6.24 and 50 µg/mL against MCF-7 (Thanomsub et al. 2007). This could be due to differences in chemical composition of the compound but most importantly on the level of purification of the compound since impure compound contains lower amount of the active compound than their pure counterparts (Sivapathasekaran et al. 2010a, b). Purification and characterization of the glycolipopeptide in this study is currently under intense investigation in our laboratory.
Re-evaluation of Surface Activities of Pseudomonas aeruginosa IKW1 Glycolipopeptide
The results of surface tension reduction showed no significant deviation from that previously reported by these authors (Ekpenyong et al. 2016). The glycolipopeptide still reduced the tensional force on the surface of fermentation medium from 71.88 to 24.61 dynes/cm at 28 ± 2 °C. This indicated that there was no significant difference (P > 0.05) in surfactant effectiveness between the data obtained in the current study and those from previous studies. Similarly, glycolipopeptide efficiency did not show any significant difference (P > 0.05) between its current critical micelle concentration of 20.69 mg/mL and the 20.8 mg/mL previously reported.
Plate 1a, ba and bb depict oil displacement and emulsification activities of the surface-active compound respectively. There was significant (P < 0.05) increment in oil displacement activity of the glycolipopeptide compound suggesting increase in the concentration of glycolipopeptide molecules obtained in this study. This activity has been reported to have a near-perfect linear relationship with surfactant concentration and is very frequently used as an indirect measurement of surface-active agent concentration (Morikawa et al. 2000). Emulsification indices (E24) significantly increased from an earlier reported 81.26% (Ekpenyong et al. 2017a) to 88.8% in kerosene (Plate 1ba) and from 82.87% (Ekpenyong et al. 2017b) to 92.57% in palm oil (Plate 1bb) at 30 °C.
Surface activities of glycolipopeptide: oil displacement activity (a) of glycolipopeptide (i) from Pseudomonas aeruginosa strain IKW1, lipopeptide (ii) from Bacillus cereus UCCM 0007 and sodium dodecyl sulfate (SDS—iii) against control (iv); emulsification activity in kerosene (b) of glycolipopeptide (i), lipopeptide (ii) and sodium dodecyl sulfate (iii); and emulsification activity in palm oil (c) of lipopeptide (i) and glycolipopeptide (ii)
Conclusion
Response surface methodology and neural network methods were compared in terms of their predictive modeling capabilities. Neural network approach showed superior predictive abilities by reason of its better MAE, MSE, Pearson’s χ2 measure and RMSE using the q-value of a volcano plot of multiple t-test analysis. The tendency to under-predict was found to reside in both techniques as evidenced in the frequency and magnitude of outliers which form the basis for large MSE values and detected by MPE. This suggests that both modeling approaches are good and reliable. The response optimizer suggested predictor variables at their optimum settings of (X1, X2, X3, X4) = (1, 1, − 1, − 1), which corresponded to actual values of temperature-32 °C, pH-7.6, agitation-130 rpm and fermentation time-66 h, with a production yield, Yp/x of 4.24, at a desirability function of 0.872. Validation of these conditions gave a glycolipopeptide concentration of 108.54 g/L and a biomass concentration of 25.49 g/L which resulted in a specific product yield, Yp/x of 4.26 g/g/DCW/h. The glycolipopeptide demonstrated commendable antimicrobial, anti-biofilm and anticancer activities especially against MCF-7 and HeLa cell lines. The compound is recommended for further studies in terms of purification, characterization and pilot-scale production for applications in treatment of chronic candidiasis by Candida dubliniensis and vaccine development for immunotherapy of breast and cervical cancers.
References
Ali N, Wang F, Xu B, Safdar B, Ullah A, Naveed M, Wang C, Rashid MT (2019) Production and application of biosurfactant produced by Bacillus licheniformis Ali5 in enhanced oil recovery and motor oil removal from contaminated sand. Molecules 24:4448. https://doi.org/10.3390/molecules24244448
Almeida DG, Soares Da Silva RC, Luna JM, Rufino RD, Santos VA, Banat IM, Sarubbo LA (2016) Biosurfactants: promising molecules for petroleum biotechnology advances. Front Microbiol 7:1718. https://doi.org/10.3389/fmicb.2016.01718
Alsaqer S, Marafi M, Banat IM, Ismail W (2018) Biosurfactant-facilitated leaching of metals from spent hydrodesulphurization catalyst. J Appl Microbiol 125:1358–1369. https://doi.org/10.1111/jam.14036
Bertrand B, Munoz-Garay C (2019) Marine antimicrobial peptides: a promising source of new generation antibiotics and other bio-active molecules. Int J Pept Res Ther 25:1441–1450. https://doi.org/10.1007/s10989-018-9789-3
Bertrand B, Martínez-Morales F, Rosas-Galván NS, Morales-Guzmán D, Trejo-Hernández MR (2018) Statistical design, a powerful tool for optimizing biosurfactant production: a review. Colloids Interfaces 2:36. https://doi.org/10.3390/colloids2030036
Bezerra KGO, Rufino RD, Luna JM, Sarrubo LA (2018) Saponins and microbial biosurfactants: potential raw materials for the formulation of cosmetics. Biotechnol Prog 34:1482–1493. https://doi.org/10.1002/btpr.2682
Bezza FA, Chirwa EMN (2017) Possible use of biosurfactants produced by microbial consortium from contaminated soil for microbially-enhanced oil recovery. CET 57:1411–1416. https://doi.org/10.3303/CET1757236
Bezza FA, Beukes M, Chirwa EMN (2015) Application of biosurfactant produced by Ochrobactrum intermedium CN3 for enhancing petroleum sludge bioremediation. Process Biochem 50:1911–1922. https://doi.org/10.1016/j.procbio.2015.07.002
Chen Y-F, Yang C-H, Chang M-S, Ciou Y-P, Huang Y-C (2010) Foam properties and detergent abilities of the saponins from Camellia oleifera. Int J Mol Sci 11:4417–4425. https://doi.org/10.3390/ijms11114417
Chouaibi M, Boussaid A, Donsi F, Ferrari G, Hamdi S (2019) Optimization of the extraction process by response surface methodology of protein isolate from defatted jujube (Zizyphus lotus L.) seeds. Int J Pept Res Ther 25:1509–1521. https://doi.org/10.1007/s10989-018-9796-4
CLSI (2019) Performance standards for antimicrobial susceptibility testing. CLSI supplement M100, 29th edn. Clinical Laboratory Standards Instituite, Wayne.
da Rocha Junior RB, Meira HM, Almeida DG, Rufino RD, Luna JM, Santos VA, Sarubbo LA (2019) Application of a low-cost biosurfactant in heavy metal remediation processes. Biodegradation 30:215–233. https://doi.org/10.1007/s10532-018-9833-1
Desai AJ, Patel KM, Desai JD (1988) Emulsifier production by Pseudomonas fluorescens during the growth on hydrocarbons. Curr Sci 57:500–501
Ebadipour N, Bagheri-Lotfabad T, Yaghmaei S, RoostaAzad R (2016) Optimization of low-cost biosurfactant production from agricultural residues through response surface methodology. Prep Biochem Biotechnol 46:30–38. https://doi.org/10.1080/10826068.2014.979204
Edet PE, Asitok AD, Ekpenyong MG, Antai SP (2018) Evaluation of the effects of nutritional and environmental parameters on production of extracellular caseinolytic protease by Stenotrophomonas acidaminiphila strain BPE4. Int J Sci 7:70–81
Ekpenyong MG, Antai SP, Asitok AD (2016) A Pseudomonas aeruginosa strain IKW1 produces an unusual polymeric surface-active compound in waste frying oil-minimal medium. Int J Sci 5:108–123
Ekpenyong M, Antai S, Asitok A, Ekpo B (2017a) Response surface modeling and optimization of major medium variables for glycolipopeptide production. Biocatal Agric Biotechnol 10:113–121. https://doi.org/10.1016/j.bcab.2017.02.015
Ekpenyong MG, Antai SP, Asitok AD, Ekpo BO (2017b) Plackett-Burman design and response surface optimization of medium trace nutrients for glycolipopeptide biosurfactant production. Iran Biomed J 21:249–260
El-Housseiny GS, Aboshanab KM, Aboulwafa MM, Hassouna NA (2019) Rhamnolipid production by a gamma ray-induced Pseudomonas aeruginosa mutant under solid state fermentation. AMB Express 9:7. https://doi.org/10.1186/s13568-018-0732-y
Franklin MJ, Nivens DE, Weadge JT, Howell PL (2011) Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel and Psl. Front Microbiol 2: 167.
Ghasemi A, Moosavi-Nasab M, Behzadnia A, Rezaei M (2018) Enhanced biosurfactant production with low-quality date syrup by Lactobacillus rhamnosus using a fed-batch fermentation. Food Sci Biotechnol 27:1137–1144. https://doi.org/10.1007/s10068-018-0366-5
Gutierrez T, Banat IM (2015) Isolation of glycoprotein bioemulsifiers produced by marine bacteria. In: McGenity T, Timmis K, Nogales B (eds) Hydrocarbon and lipid microbiology protocols. Springer, Berlin, pp 61–74
Hippolyte MT, Augustin M, Hervé TM, Robert N, Devappa S (2018) Application of response surface methodology to improve the production of antimicrobial biosurfactants by Lactobacillus paracasei subsp. tolerans N2 using sugar cane molasses as substrate. Bioresour Bioprocess 5:48. https://doi.org/10.1186/s40643-018-0234-4
Hisatsuka K, Nakahara T, Yamada K (1972) Protein-like activator for n-alkane oxidation by Pseudomonas aeruginosa S7B1. Agric Biol Chem 36:1361–1369. https://doi.org/10.1271/bbb1961.36.1361
Hoskova M, Jezdik R, Schreiberova O, Chudoba J, Sir M, Cejkova A, Masak J, Jirku V, Rezanka T (2015) Structural and physiochemical characterization of rhamnolipids produced by Acinetobacter calcoaceticus, Enterobacter asburiae and Pseudomonas aeruginosa in single strain and mixed cultures. J Biotechnol 193:45–51. https://doi.org/10.1016/j.jbiotech.2014.11.014
Hu F, Liu Y, Li S (2019) Rational strain improvement for surfactin production: enhancing the yield and generating novel structures. Microb Cell Fact 18:42. https://doi.org/10.1186/s12934-019-1089-x
Ilori MO, Amund DI (2001) Production of a peptidoglycolipid bioemulsifier from Pseudomonas aeruginosa grown on hydrocarbon. Z Naturforsch C 56:547–552. https://doi.org/10.1515/znc-2001-7-812
Javee A, Karuppan R, Subramani N (2020) Bioactive glycolipid biosurfactant from seaweed Sargassum myriocystum-associated bacteria Streptomyces sp. SNJASM6. Biocatal Agric Biotechnol 23:101505. https://doi.org/10.1016/j.bcab.2020.101505
Jimoh AA, Lin J (2019) Biosurfactant: A new frontier for greener technology and environmental sustainability. Ecotoxicol Environ Saf 184:109607. https://doi.org/10.1016/j.ecoenv.2019.109607
Karlapudi AP, Venkateswarulu TC, Srirama K, Kota RK, Mikkili I, Kodali VP (2020) Evaluation of anti-cancer, anti-microbial and anti-biofilm potential of biosurfactant extracted from an Acinetobacter M6 strain. J King Saud Univ Sci 32:223–227. https://doi.org/10.1016/j.jksus.2018.04.007
Karri RR, Tanzifi M, Yaraki MT, Sahu JN (2018) Optimization and modeling of methyl orange adsorption onto polyaniline nano-adsorbent through response surface methodology and differential evolution embedded neural network. J Environ Manag 223:517–529. https://doi.org/10.1016/j.jenvman.2018.06.027
Kim J-S, Lee I-K, Yun BS (2018) Pullusurfactans A-E, new biosurfactants produced by Aureobasidium pullulans A11211-4-57 from a fleabane, Erigeron annus (L.) pers. J Antibiot 71:920–926. https://doi.org/10.1038/s41429-018-0089-0
Kiran GS, Hema TA, Gandhimathi R, Selvin J, Thomas TA, Ravji TR, Natarajaseenivasan K (2009) Optimization and production of a biosurfactant from the sponge-associated marine fungus Aspergillus ustus MSF3. Colloids Surf B 73:250–256. https://doi.org/10.1016/j.colsurfb.2009.05.025
Koronelli TV, Komarova TI, Denisov LV (1983) Chemical composition and role of Pseudomonas aeruginosa peptidoglycolipid in hydrocarbon assimilation. Mikrobiologiia 52:767–777
Kuiper I, Lagendijk L, Pickford R, Derrick JP, Lamers GEM, Thomas-Oates JE, Lugtenberg BJJ, Bloemberg GV (2004) Characterization of two Pseudomonas putida lipopeptide biosurfactants, Putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Mol Microbiol 51:97–113. https://doi.org/10.1046/j.1365-2958.2003.03751.x
Mabrouk MEM, Youssif EM, Sabry SA (2014) Biosurfcatant production by a newly isolated soft coral-associated marine Bacillus sp.E34: Statistical optimization and characterization. Life Sci J 11:756–768
Martinez-Toledo A, Rodriguez-Vazquez R (2013) In situ biosurfactant production and hydrocarbon removal by Pseudomonas putida CB-100 in bioaugmented and biostimulated oil-contaminated soil. Braz J Microbiol 44:595–605. https://doi.org/10.1590/S1517-83822013000200040
McDonald DM, Byrne SN, Payne RJ (2015) Synthetic self-adjuvanting glycopeptide cancer vaccines. Front Chem 3:60. https://doi.org/10.3389/fchem.2015.00060
Morais IMC, Cordeiro AL, Teixeira GS, Domingues VS, Nardi RMD, Monteiro AS, Alves RJ, Siqueira EP, Santos VL (2017) Biological and physicochemical properties of biosurfactants produced by Lactobacillus jensenii P6A and Lactobacillus gasseri P65. Microb Cell Fact 16:155. https://doi.org/10.1186/s12934-017-0769-7
Morikawa M, Hirata Y, Imanaka T (2000) A study on the structure-function relationship of lipopeptides biosurfactants. Biochim Biophys Acta 1488:211–218
Myers RH, Montgomery DC (2002) Response surface methodology: product and process optimization using designed experiments. Wiley, New York
Nicolo MS, Cambria MG, Impallomeni G, Rizzo MG, Pellicorio C, Ballistreri A, Guglielmino SPP (2017) Carbon source effects on the mono/dirhamnolipid ratio produced by Pseudomonas aeruginosa L05, a new human respiratory isolate. N Biotechnol 39:36–41. https://doi.org/10.1016/j.nbt.2017.05.013
Pal MP, Vaidya BK, Desai KM, Joshi RM, Nene SN, Kulkarni BD (2009) Media optimization for biosurfactant production by Rhodococcus erythropolis MTCC 2794: artificial intelligence versus a statistical approach. J Ind Microbiol Biotechnol 36:747–756. https://doi.org/10.1007/s10295-009-0547-6
Patel KA, Brahmbhatt PK (2016) A comparative study of the RSM and ANN models for predicting surface roughness in roller burnishing. Procedia Technol 23:391–397. https://doi.org/10.1016/j.protcy.2016.03.042
Perfumo A, Rudden M, Marchant R, Banat IM (2017) Biodiversity of biosurfactants and roles in enhancing the (bio) availability of hydrophobic substrates. In: Krell T (ed) Cellular ecophysiology of microbe. Handbook of hydrocarbon and lipid microbiology. Springer, Cham, pp 1–29
Pilkington JL, Preston C, Gomes RL (2014) Comparison of response surface methodology (RSM) and artificial neural networks (ANN) towards efficient extraction of artemisinin from Artemisia annua. Ind Crops Prod 58:15–24. https://doi.org/10.1016/j.indcrop.2014.03.016
Rani M, Weadge JT, Jabaji S (2020) Isolation and characterization of biosurfactant-producing bacteria from oil well batteries with antimicrobial activities against food-borne and plant pathogens. Front Microbiol 11:64. https://doi.org/10.3389/fmicb.2020.00064
Renaudet O, Dasgupta G, Bettahi I, Shi A, Nesburn AB, Dumy P, BenMohamed L (2010) Linear and branched glyco-lipopeptide vaccines follow distinct cross-presentation pathways and generate different magnitudes of antitumor immunity. PLoS ONE 5:e11216. https://doi.org/10.1371/journal.pone.0011216
Rodrigues L, Teixeira J, Oliveira R, Van der Mei HC (2006) Response surface optimization of the medium components for the production of biosurfactants by probiotic bacteria. Process Biochem 41:1–10. https://doi.org/10.1016/j.procbio.2005.01.030
Rodrigues AC, Fontao AI, Coelho A, Leal M, Soares da Silva FAG, Wan Y, Dourado F, Gama M (2019) Response surface statistical optimization of bacterial nanocellulose fermentation in static culture using a low-cost medium. New Biotechnol 49:19–27. https://doi.org/10.1016/j.nbt.2018.12.002
Sampaio FC, Teles de Faria J, Silva GD, Gonçalves RM, Pitangui CG, Casazza AA, Arni SA, Converti A (2017) Comparison of response surface methodology and artificial neural network for modeling xylose-to-xylitol bioconversion. Chem Eng Technol 40:122–129. https://doi.org/10.1002/ceat.201600066
Santos BF, Ponezi AN, Fileti AMF (2017) Development of artificial intelligence models to monitor biosurfactant concentration in real-time using waste as substrate in bioreactor through fermentation by Bacillus subtilis. Chem Eng Trans 57:1009–1014. https://doi.org/10.3303/CET1757169
Satpute SK, Kulkarni GR, Banpurkar AG, Banat IM, Mone NS, Patil RH, Cameotra SS (2016) Biosurfactnat/s from Lactobacilli species: properties, challenges and potential biomedical applications. J Basic Microbiol 56:1–19. https://doi.org/10.1002/jobm.201600143
Satpute SK, Mone NS, Das P, Banpurkar AG, Banat IM (2018a) Lactobacillus acidophilus derived biosurfactant as a biofilm inhibitor: a promising investigation using microfluidic approach. Appl Sci 8:1555. https://doi.org/10.3390/app8091555
Satpute SK, Zinjarde SS, Banat IM (2018b) Recent updates on biosurfactants in food industry. In: Sharma D, Saharan BS (eds) Microbial Cell Factories. CRC Press, Boca Raton, pp 1–20
Sivapathasekaran C, Das P, Mukherjee S, Saravanakumar J, Mandal M, Sen R (2010a) Marine bacterium derived lipopeptides: characterization and cytotoxic activity against cancer cell lines. Int J Pept Res Ther 16:215–222. https://doi.org/10.1007/s10989-010-9212-1
Sivapathasekaran C, Mukherjee S, Ray A, Gupta A, Sen R (2010b) Artificial neural network modeling and genetic algorithm based medium optimization for the improved production of marine biosurfactant. Bioresour Technol 101:2884–2887. https://doi.org/10.1016/j.biortech.2009.09.093
Terziyski I, Alexandrova L, Stoineva I, Christova N, Todorov R, Cohen R (2014) Foam and wetting films from rhamnolipids produced by Pseudomonas aeruginosa BN10. Colloids Surf A 460:299–305. https://doi.org/10.1016/j.colsurfa.2013.12.075
Thanomsub B, Pumeechockchai W, Limtrakul A, Arunrattiyakorn P, Petchleelaha W, Nitoda T, Kanzaki H (2007) Chemical structures and biological activities of rhamnolipid produced by Pseudomonas aeruginosa B189 isolated from milk factory waste. Bioresour Technol 98:1149–1153. https://doi.org/10.1016/j.biortech.2005.10.029
Thavasi R, Jayalakshmi S, Thangavel B, Banat I (2008) Production and characterization of a glycolipid biosurfactant from Bacillus megaterium using economically cheaper sources. World J Microbiol Biotechnol 24:917–925. https://doi.org/10.1007/s11274-007-9609-y
Thavasi RT, Jayalakshmi S, Banat IM (2011a) Effect of biosurfactant and fertilizer on biodegradation of crude oil by marine isolates of Bacillus megaterium, Corynebacterium kutscheri and Pseudomonas aeruginosa. Bioresour Technol 102:772–778. https://doi.org/10.1016/j.biortech.2010.08.099
Thavasi R, Subramanyam-Nambaru VRM, Jayalakshmi S, Balasubramanian T, Banat IM (2011b) Biosurfactant production by Pseudomonas aeruginosa from renewable resources. Indian J Microbiol 51:30–36. https://doi.org/10.1007/s12088-011-0076-7
Truppo MD (2017) Biocatalysis in the pharmaceutical industries: the need for speed. ACS Med Chem Lett 8:476–480. https://doi.org/10.1021/acsmedchemlett.7b00114
Vecino X, Rodriguez-Lopes L, Ferreira D, Cruz JM, Moldes AB, Rodrigues LR (2018) Bioactivity of glycolipopeptide cell-bound biosurfactants against skin pathogens. Int J Biol Macromol 109:971–979. https://doi.org/10.1016/j.ijbiomac.2017.11.088
Wachtmeister J, Rother D (2017) Recent advances in whole cell biocatalysis techniques bridging from investigative to industrial scale. Curr Opin Biotechnol 42:169–177. https://doi.org/10.1016/j.copbio.2016.05.005
Wu Y-S, Ngai S-C, Goh B-H, Chan K-G, Lee L-H, Chuah L-H (2017) Anti-cancer activities of surfactin and potential application of nanotechnology assisted surfactin delivery. Front Pharmacol 8:781. https://doi.org/10.3389/fphar.2017.00761
Acknowledgements
This work was supported by government of Cross River State, Nigeria (Grant No.: CRS/SB/P/149/30). The authors are also grateful to Dr. Samuel Ekpata for his financial assistance and Prof. O. E. Fagade of the University of Ibadan for facilitating glycolipopeptide analyses and identification.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there was no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ekpenyong, M., Asitok, A., Antai, S. et al. Statistical and Artificial Neural Network Approaches to Modeling and Optimization of Fermentation Conditions for Production of a Surface/Bioactive Glyco-lipo-peptide. Int J Pept Res Ther 27, 475–495 (2021). https://doi.org/10.1007/s10989-020-10094-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-020-10094-8