Abstract
We have analysed the response of cells on a bed of micro-posts idealized as a Winkler foundation using a homeostatic mechanics framework. The framework enables quantitative estimates of the stochastic response of cells along with the coupled analysis of cell spreading, contractility and mechano-sensitivity. In particular the model is shown to accurately predict that: (i) the extent of cell spreading, actin polymerisation as well as the traction forces that cells exert increase with increasing stiffness of the foundation; (ii) the traction forces that cells exert are primarily concentrated along the cell periphery; and (iii) while the total tractions increase with increasing cell area the average tractions are reasonably independent of cell area, i.e. for a given substrate stiffness, the average tractions that are normalized by cell area do not vary strongly with cell size. These results thus suggest that the increased foundation stiffness causes both the cell area and the average tractions that the cells exert to increase through higher levels of stress-fibre polymerization rather than the enhanced total tractions being directly linked through causation to the larger cell areas. A defining feature of the model is that its predictions are statistical in the form of probability distributions of observables such as the traction forces and cell area. In contrast, most existing models present solutions to specific boundary value problems where the cell morphology is imposed a priori. In particular, in line with observations we predict that the diversity of cell shapes, sizes and measured traction forces increase with increasing foundation stiffness. The homeostatic mechanics framework thus suggests that the diversity of observations in in vitro experiments is inherent to the homeostatic equilibrium of cells rather than being a result of experimental errors.
Similar content being viewed by others
1 Introduction
Living cells sense and respond to their mechanical, chemical and topological environments. For example, Engler et al. [1] have shown that cell spreading is strongly affected by substrate stiffness with the projected cell area increasing with increasing substrate stiffness. Similar observations showing that cell shape as characterized by aspect ratio is also dependent on substrate stiffness [2] have also been reported. This dependence of direct visual observables to substrate stiffness is linked to the forces that cells exert on the substrate with the total traction forces also increasing with increasing substrate stiffness [3, 4]. However, while the statistics of these behaviours are highly reproducible, cells display a fluctuating response that results in a diversity of observables in nominally identical tests. Importantly, this experimental variability is not only a function of the cell type but also a function of the environment with the standard deviation in all the quantities mentioned above decreasing with decreasing substrate stiffness. The variability in direct observables such as cell shape, area and cytoskeletal protein arrangements is also linked to other critical cell functionality. In particular, mechanical, geometric and topological cues direct the differentiation of mesenchymal stem cells (MSCs) [5,6,7]. However, this behaviour is stochastic such that MSCs differentiate mainly but not exclusively into bone cells when cultured on stiff substrates while the probability to differentiate into neuronal cells increases on soft substrates [5]. Thus, the observed responses of cells are always characterised in terms of statistics rather than unique outcomes. A mechanistic understanding of this stochastic behaviour of cells will have far-reaching implications in aiding the interpretation of a wide range of cell functionalities and responses.
Cytoskeletal tension plays an essential role in the way cells sense and respond to their environment with the above discussed sensitivities suppressed when inhibitors such as cytochalasin D are added to diminish myosin contractility [5, 8, 9]. This understanding has spurred extensive experimental activity to measure the traction forces that cells exert. The main approaches include traction force microscopy [10, 11] and using deformable micro-post arrays [12,13,14,15]. The key conclusions from these studies are: (i) the so-called total traction forces rise with increasing substrate stiffness; and (ii) cells spread to a greater extent on stiffer substrates. In fact, both these processes are coupled in the sense that cells undergo remodelling and reorganize their cytoskeleton in response to their mechanical environment, with the cytoskeletal structure controlling both cell shape/area as well as the traction forces that are generated.
A number of modelling approaches have been developed to model the mechano-sensitive response of cells with the aim of using models to try and interpret the traction force measurements. Early attempts included modelling the cytoskeleton as an interlinked structure of passive filaments [16] or modelling contractility as a thermal contraction [17]. Deshpande et al. [18] introduced a bio-chemo-mechanical model for cell contractility that subsequently has been adapted in a number of studies [19,20,21]. These models rationalised a number of the observations of cells on elastic substrates but inherently solved a boundary value problem such that the size and shape of the cell was specified a priori. In reality, contractility and the shape that cells adopt by spreading are interlinked with both, in turn, depending on environmental parameters such as substrate stiffness and ligand density [1]. Comprehensive intricate coupling among cell configuration, traction forces and the elastic environment is missing in all these modelling approaches. Moreover, they necessarily give only deterministic predictions for the response of cells while the measured responses are clearly stochastic.
The key assumption in all the models mentioned above is that they presume, either implicitly or explicitly, that a system comprising the cell and the substrate attains equilibrium at its minimum value of Gibbs free-energy. However, such a system that excludes the nutrient bath surrounding the cell is an open system with the cell exchanging (chemical) species with the nutrient bath (Fig. 1a). In fact, thermodynamic equilibrium of this open system is achieved when the chemical potentials of all mobile species within the cell and nutrient bath equalise but living cells never achieve such an equilibrium state (e.g. all living cells maintain a resting potential between the cell and the surrounding nutrient bath by actively regulating the concentration of various ions within the cell). Hence, cells are inherently in a non-equilibrium state from a conventional thermodynamic perspective. Shishvan et al. [22] have recently proposed a homeostatic ensemble to describe the dynamic equilibrium of cells in an attempt to resolve this critical limitation in all previous modelling approaches. This framework therefore inherently allows us to make quantitative predictions of the stochastic response of cells with cell shape and size no longer imposed via a boundary value problem but rather the distribution of configurations that cells adopt being an outcome of the homeostatic equilibrium in the given environment.
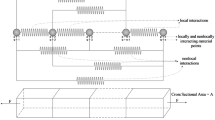
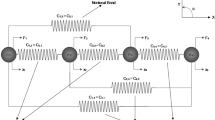
a Sketch of a cell on a bed of micro-posts. b Sketch of the cell on a Winkler foundation used to approximate the bed of micro-posts. In a and b the nutrient bath has been illustrated and a small selection of the species being exchanged between the cell and the bath are labelled. c The two-dimensional (2D) approximation of a cell on the Winkler foundation analysed in this study
The homeostatic statistical mechanics description for cells [22] has already been shown to successfully capture a range of observations for smooth muscle cells seeded on elastic substrates [22, 23] and for myofibroblasts seeded on substrates micropatterned with stripes of fibronectin [24, 25] as well as for the differentiation of hMSCs in response to a range of environmental cues including stiffness of substrates and sizes of adhesive islands [26]. These give us confidence in utilizing the homeostatic mechanics framework to investigate the response of cells on a dense array of micro-posts.
1.1 Cells on a dense array of micro-posts
Here we consider the problem of a cell on a bed of micro-posts arranged in a cubic pattern with spacing \(L\), as sketched in Fig. 1a. The posts have a height \(H\) and in line with most experimental studies [12,13,14,15] are assumed to have a circular cross-section of radius \(a\), as sketched in Fig. 1a. The bending stiffness \(S\) of the posts is then
where \(E\) is the Young’s modulus of micro-posts. If a representative spread dimension \(R\) of the cell in the \({x}_{1}-{x}_{2}\) plane is much greater than the post spacing, i.e. \(R\gg L\), then it suffices to approximate the bed of posts as an effective Winkler foundation with smeared-out isotropic stiffness \(\kappa =S/{L}^{2}\) in the \({x}_{1}-{x}_{2}\) plane (Fig. 1b). In this study we shall analyse the behaviour of smooth muscle cells (SMCs) on a dense array of micro-posts using this Winkler approximation. We emphasize that the Winkler-foundation approximation implies that the precise post geometry does not enter the analysis explicitly. Rather the analysis only depends on the Winkler stiffness \(\kappa \).
The outline of the paper is as follows. We first briefly review the model of Vigliotti et al. [27] as modified in [22] to calculate the free-energy of a system comprising the cell and foundation. Living cells, however, do not attain thermodynamic equilibrium and thus do not equilibrate at a minimum free-energy state. Next, we briefly describe the homeostatic mechanics framework [22] to account for the dynamical equilibrium that living cells attain. Finally, we present predictions using the homeostatic mechanics framework for the responses of cells on arrays of micro-posts in an attempt to identify correlations or the lack of them in the dependences of cell area, cell shape and traction forces on the stiffness of the micro-post arrays.
2 Free-energy of a cell on a dense array of posts
Consider a cell in a nutrient bath on a bed of micro-posts (approximated as a Winkler foundation), as sketched in Fig. 1b. We define a system comprising the cell and foundation but absent the nutrient bath which is considered as the environment. This of course has the advantage that it is not then required to model the nutrient bath but rather only model the parts of the experimental setup on which observations are being directly conducted. The state of the system changes as the cell moves, spreads and changes shape on the foundation and here we shall give a prescription to calculate the free-energy of the cell in a specific configuration \((j)\) where the connections of material points on the cell membrane to the foundation surface are specified (with the remainder of the cell surface and foundation assumed to be traction-free). In broad terms the adhesion of the cell membrane to the foundation specifies the shape of the cell and subsequently we shall refer to each such configuration as a morphological microstate \((j)\) of the system.
2.1 The model for the Gibbs free-energy
With the system comprising of the cell and the foundation within a constant temperature and pressure nutrient bath, the Gibbs free-energy \({\mathcal{G}}^{(j)}\) of the system in morphological microstate \((j)\) is given by
where \(f\) is the specific Helmholtz free-energy of the cell and \(\psi \) the strain energy density of the foundation. We emphasize that the analysis presented here is for the system under atmospheric pressure conditions and thus without loss of generality we set \(p=0\) (i.e. use gauge pressure). Thus, a pressure term does not appear in (2). The equilibrium free-energy \({G}^{(j)}\) is then the value of \({\mathcal{G}}^{(j)}\) at \({d\mathcal{G}}^{(j)}=0\). Here, we briefly describe the model for the calculation of \({G}^{(j)}\). In the following, for the sake of notational brevity, we shall drop the superscript \((j)\) that denotes the morphological microstate as the entire discussion refers to a single morphological microstate.
With the cell exerting a spatial distribution of tractions \({T}_{1}({x}_{i})\) and \({T}_{2}({x}_{i})\) on the foundation, the strain energy density \(\psi \) is given by
The total foundation strain energy
where the spatial integration is carried out over the current cell area \(A\). We now proceed to summarise the model of Vigliotti et al. [27] for the cell as modified in [22] for a non-dilute concentration of stress-fibres. Here, we restrict ourselves to a two-dimensional (2D) approximation of the cell (Fig. 1c) such that the cell lies in the \({x}_{1}-{x}_{2}\) plane with no variation of properties in the through thickness \({x}_{3}\)-direction.
The Vigliotti et al. [27] model assumes only two elements within the cell: (i) a passive elastic contribution from elements such as the cell membrane, intermediate filaments and microtubules and (ii) contractile acto-myosin stress-fibres that are modelled explicitly. Consider a 2D cell of thickness \({b}_{0}\) and volume \({V}_{0}\) in its elastic resting state. The representative volume element (RVE) of the stress-fibres within the cell in this resting configuration is assumed to be a cylinder of volume \({V}_{{\text{R}}}=\pi {b}_{0}{\left(\frac{{n}^{{\text{R}}}{\ell}_{0}}{2}\right)}^{2}\) where \({\ell}_{0}\) is the length of a stress-fibre functional unit in its ground-state and \({n}^{{\text{R}}}\) is the number of these ground-state functional units within this reference RVE. The total number of functional unit packets within the cell is \({N}_{0}^{{\text{T}}}\) and we introduce \({N}_{0}={N}_{0}^{{\text{T}}}{V}_{{\text{R}}}/{V}_{0}\) as the average number of functional unit packets available per RVE; \({N}_{0}\) shall serve as a useful normalisation parameter. The state of the stress-fibres at location \({x}_{i}\) within the cell is described by their angular concentration \(\eta ({x}_{i},\phi)\), and there are \(n({x}_{i},\phi )\) functional units in series along the length of each stress-fiber in the RVE. Here, \(\phi \) is the angle with respect to the \({x}_{1}\) direction (Fig. 1c). Vigliotti et al. [27] argue that an applied stretch is shared equally among all subunits, so that the strain within each functional unit, \(\tilde{\varepsilon }_{{\text{nom}}}\), is initially equal to the nominal strain \({\varepsilon }_{{\text{nom}}}({x}_{i},\phi )\) in direction \(\phi \). Subsequent addition or removal of subunits modifies the subunit stretch proportionally so that, at steady-state, the number \({n}^{{\text{ss}}}\) of functional units within the stress fibers is given by
where \(\tilde{\varepsilon}_{{\text{nom}}}^{{\text{ss}}}\) is the strain at steady-state within a functional unit of the stress-fibres. It now remains to specify the steady-state angular concentration of the stress-fibres. The chemical potential of the functional units within the stress-fibres is given by [22]
where k and T are Boltzmann constant and temperature, respectively, and the normalized concentration of the unbound stress fiber proteins is given by \({\hat{N}}_{{\text{u}}}\equiv {N}_{{\text{u}}}/{N}_{0}\). With \(\hat{\eta }\equiv \eta {n}^{{\text{R}}}/{N}_{0}\), the maximum normalised value of \(\hat{\eta }\), denoted by \({\hat{\eta }}_{{\text{max}}}\), corresponds to full occupancy of all available sites for stress-fibres. Here, \({\hat{N}}_{{\text{L}}}\) is the number of lattice sites available to unbound proteins. The enthalpy \({\mu }_{{\text{b}}}^{{\text{ss}}}\) of \({n}^{{\text{R}}}\) bound functional units at steady-state is given in terms of the isometric stress-fibre stress \({\sigma }_{{\text{max}}}\) and the internal energy \({\mu }_{{\text{b0}}}\) as
where \(\Omega \) is the volume of \({n}^{{\text{R}}}\) functional units. By contrast, the chemical potential of the unbound proteins is independent of stress and given in terms of the internal energy \({\mu }_{{\text{u}}}\) as
For a fixed configuration of the 2D cell (i.e. a fixed strain distribution \({\varepsilon }_{{\text{nom}}}({x}_{i},\phi )\)), the contribution to the specific Helmholtz free-energy of the cell, \(f\), from the stress-fibre cytoskeleton follows as
where \({\rho }_{0}\equiv {N}_{0}/{V}_{{\text{R}}}\) is the number of protein packets per unit reference volume available to form functional units in the cell. However, we cannot yet evaluate \({f}_{{\text{cyto}}}\) as \({\hat{N}}_{{\text{u}}}({x}_{i})\) and \(\hat{\eta }({x}_{i},\phi )\) are unknown. These will follow from the chemical equilibrium of the cell as will be discussed in Sect. 2.2.
The total stress \({\varSigma }_{ij}\) within the cell includes contributions from the passive elasticity provided mainly by the intermediate filaments of the cytoskeleton attached to the nuclear and plasma membranes and the microtubules as well as the active contractile stresses of the stress-fibres. Thus, in line with the existing literature on active stress mechanics, we write the total Cauchy stress in an additive decomposition as
where \({\sigma }_{ij}\) and \({\sigma }_{ij}^{{\text{p}}}\) are the active and passive Cauchy stresses, respectively. In the 2D setting with the cell lying in the \({x}_{1}-{x}_{2}\) plane, the active stress is given in terms of the volume fraction \({\mathcal{F}}_{0}\) of the stress-fibre proteins as
where \({\phi }^{*}\) is the angle of the stress-fibre measured with respect to \({x}_{i}\) and is related to \(\phi \) by the rotation with respect to the undeformed configuration. The passive elasticity in the 2D setting is given by a 2D specialization of the Ogden [28] hyperelastic model as derived in [22]. The strain energy density function of this 2D Ogden model is
where \({\lambda }_{I}\) and \({\lambda }_{II}\) are the principal stretches, \(\mu \) and \({\rm K}\) are the shear modulus and in-plane bulk modulus, respectively, and \(m\) is a material constant governing the non-linearity of the deviatoric elastic response. Moreover, since the cell is assumed to be incompressible we set the principal stretch in the \({x}_{3}\)-direction \({\lambda }_{III}=1/{(\lambda }_{I}{\lambda }_{II})\). The (passive) Cauchy stress then follows as \({\sigma }_{ij}^{{\text{p}}}{p}_{j}^{(k)}={\sigma }_{k}^{{\text{p}}}{p}_{i}^{(k)}\) in terms of the principal (passive) Cauchy stresses \({\sigma }_{k}^{{\text{p}}}\equiv {\lambda }_{k}\partial {\varPhi }_{{\text{elas}}}/\partial {\lambda }_{k}\) and the unit vectors \({p}_{j}^{(k)} \,(k=I, II)\) in the principal directions. The total specific Helmholtz free-energy of the cell is then \({f}=f_{{\text{cyto}}}+{\varPhi }_{{\text{elas}}}\).
We emphasize here that the constitutive formulation differs considerably from the formulations employed for the inelastic deformation of non-active materials. In such non-active materials, a multiplicative decomposition of the deformation gradient in terms of the elastic and inelastic parts is assumed. By contrast, for the active behaviour of cells we assume that there is no inelastic deformation with the total deformation gradient equal to the elastic deformation gradient. However, an active stress resulting from the stress fibres is assumed to act in parallel to the passive elastic stress.
2.2 Equilibrium of the morphological microstate
Shishvan et al. [22] have shown that equilibrium of a morphological microstate reduces to two conditions: (i) mechanical equilibrium with \({\varSigma }_{ij,j}=0\) throughout the system and (ii) chemical equilibrium such that \({\chi }_{{\text{u}}}({x}_{i})={\chi }_{{\text{b}}}({x}_{i},\phi )={\text{constant}}\), i.e. the chemical potentials of bound and unbound stress-fibre proteins are equal throughout the cell. The condition \({\chi }_{{\text{u}}}={\chi }_{{\text{b}}}\) implies that \(\hat{\eta }({x}_{i},\phi )\) is given in terms of \({\hat{N}}_{{\text{u}}}\) by
and \({\hat{N}}_{{\text{u}}}\) follows from the conservation of stress-fibre proteins throughout the cell, viz.
Knowing \({\hat{N}}_{{\text{u}}}\) and \(\hat{\eta }({x}_{i},\phi )\), the stress \({\varSigma }_{ij}\) can now be evaluated and these stresses within the system (i.e. cell and foundation) need to satisfy mechanical equilibrium, i.e. \({\varSigma }_{ij,j}=0\). In this case, the mechanical equilibrium condition is readily satisfied as the stress field \({\varSigma }_{ij}\) within the cell is equilibrated by a traction field \({T}_{i}\) exerted by the foundation on the cell such that \(b{\varSigma }_{ij,j}=-{T}_{i}\), where \(b({x}_{i})\) is the thickness of the cell in the current configuration. Tractions calculated in this manner are then directly used in Eqs. (3) and (4) to obtain the foundation elastic energy.
The equilibrium value of \(\mathcal{G}\) denoted by \(G\) is then given as \(G={F}_{{\text{cell}}}+{F}_{{\text{foun}}}\) where
and \({F}_{{\text{foun}}}\) is specified by Eq. (4). Here, \({\chi }_{u}\) is given by Eq. (8) with the equilibrium value of \({\hat{N}}_{{\text{u}}}\) obtained from Eq. (14). For the purposes of further discussion, we define the equilibrium value \({{F}_{{\text{cyto}}}\equiv \rho }_{0}{V}_{0}{\chi }_{{\text{u}}}\) as the cytoskeletal free-energy of the cell and \({F}_{{\text{passive}}}\equiv {\int }_{{V}_{{\text{cell}}}}{\varPhi }_{{\text{elas}}} dV\) as the passive elastic energy of the cell.
The free-energy \(G\) can be decomposed as \(G=\Upsilon\) + \({\Upsilon}_{0}\), where \({\Upsilon}_{0}={\rho }_{0}{V}_{0}\left[{\mu }_{{\text{u}}}/{n}^{{\text{R}}}-kT \ln\left(\pi {\hat{N}}_{{\text{L}}}\right)\right]\) is independent of the morphological microstate. It is thus natural to subtract out \({\Upsilon}_{0}\) and define a normalised free-energy as
where \({G}_{{\text{S}}}\) is the equilibrium free-energy of a free-standing cell (i.e. a cell in suspension with traction-free surfaces). Analogously, we define the normalised passive and cytoskeletal free-energies of the cell as
and
respectively. We shall present all energies in these normalised forms.
2.3 Model parameters
The simulations were performed with the parameters taken from Shishvan et al. [22] and are representative of smooth muscle cells (SMCs). All simulations are reported at a reference thermodynamic temperature \(T={T}_{0}\), where \({T}_{0}=310\, {\text{K}}\). The passive elastic parameters of the cell are taken to be \(\mu =1.67\, {\text{kPa}}\), \({\rm K}=35\, {\text{kPa}}\) and \(m=6\). For SMCs, the maximum contractile stress \({\sigma }_{{\text{max}}}=240\, {\text{kPa}}\) consistent with a wide range of measurements on muscle fibres [29] and the density of stress-fibre proteins was taken as \({\rho }_{0}=3\times {10}^{6}\, \upmu {{\text{m}}}^{-3}\) with the volume fraction of stress-fibre proteins \({\mathcal{F}}_{0}=0.032\). Following Vigliotti et al. [27], we assume that the steady-state functional unit strain \(\tilde{\varepsilon}_{{\text{nom}}}^{{\text{ss}}}=0.35\) with \({\mu }_{{\text{b0}}}-{\mu }_{{\text{u}}}=2.3 k{T}_{0}\) and \(\Omega ={10}^{-7.1} \,\upmu {{\text{m}}}^{3}\). The maximum angular stress fibre concentrations was set to be \({\hat{\eta }}_{{\text{max}}}=1\) based on the assumption that the local density of bound stress-fibre proteins cannot exceed \({\rho }_{0}\). All results are presented for a cell that is assumed to be circular with a radius \({R}_{0}\) and thickness \({b}_{0}\) in its undeformed state with \({b}_{0}/{R}_{0}=0.2\). Since results are presented with energies normalised as per (16)–(18), the quantities \({\mu }_{{\text{u}}}, {n}^{{\text{R}}}\) and \({\hat{N}}_{{\text{L}}}\) do not need to be explicitly specified. With these parameters, the equilibrium free-energy of the free-standing cell is \({(G}_{{\text{S}}}-{\Upsilon}_{0})/({V}_{0}k{T}_{0})\approx -5.6\times {10}^{6} \,\upmu {{\text{m}}}^{-3}\) where \({V}_{0}=\pi {R}_{0}^{2}{b}_{0}\) is the cell volume. In this free-standing state the equilibrium configuration of the cell is a spatially uniform cylinder with radius \(\sim 0.92{R}_{0}\).
2.4 Competition between elastic and cytoskeletal free-energy
One of the key features of the constitutive model of Vigliotti et al. [27] is the competition between the elastic free-energy \({F}_{{\text{passive}}}\) and the cytoskeletal free-energy \({F}_{{\text{cyto}}}\) of the cell that sets the free-energy of the cell. To illustrate this competition, we consider the highly simplified problem of a circular cell on a rigid foundation and constrain ourselves to morphological microstates wherein the strain distribution within the cell is spatially uniform. We emphasize here that this is an unrealistic restriction of the phase-space of morphological microstates that the cell will attain and this restriction is relaxed when the homeostatic mechanics is introduced in Sect. 3. However, for the purposes of illustrating the basic physics of the free-energy model we present this restrictive analysis here in which a morphological microstate is described by one scalar variable, e.g. the area \(A\) of the cell.
For the case of a cell on a rigid foundation, there is no contribution to the Gibbs free-energy of the system from the foundation and so \(G={F}_{{\text{cell}}}\). The normalised free-energy of the system \(\hat{G}\) is plotted in Fig. 2a as a function of the normalised cell area \(\hat{A}\equiv A/{A}_{0}\), where \({A}_{0}\) is the area of the undeformed cell. There is a clear minimum of \(\hat{G}\) at \({\hat{A}}_{{\text{opt}}}\approx 1.44\). To understand this minimum, the variations of the free-energies \({\hat{F}}_{{\text{passive}}}\) and \({\hat{F}}_{{\text{cyto}}}\) with \(\hat{A}\) are also included in Fig. 2a. The elastic energy increases with increasing \(|\hat{A}-1|\) as the cell is strained away from its undeformed configuration. By contrast, \({\hat{F}}_{{\text{cyto}}}\) decreases monotonically with increasing \(\hat{A}\). This competition between \({\hat{F}}_{{\text{passive}}}\) and \({\hat{F}}_{{\text{cyto}}}\) with varying \(\hat{A}\) gives rise to the minimum in \(\hat{G}\). Assuming that the cell is evolving towards a minimum free-energy configuration (this is clearly not true and we shall show in Sect. 3 that low Gibbs free-energy states are just more probable), we can say that the stress-fibre cytoskeleton drives cell spreading. This is consistent with a large number of observations [8, 9] that indicate that inhibiting stress-fibres via reagents such as cytochalasin D and blebbistatin reduces cell spreading. In fact, the model predicts reduced spreading as the available stress-fibre proteins are reduced and this is illustrated in Fig. 2b where we show the dependence of \({\hat{A}}_{{\text{opt}}}\) on \({\rho }_{0}\) keeping all other parameters fixed. With decreasing \({\rho }_{0}\) the area \({\hat{A}}_{{\text{opt}}}\) at which the Gibbs free-energy is minimized becomes closer to the undeformed state with \({\hat{A}}_{{\text{opt}}}=1\).
a The normalised Gibbs free-energy \(\hat{G}\), passive elastic energy \({\hat{F}}_{{\text{passive}}}\) and the cytoskeletal free-energy \({\hat{F}}_{{\text{cyto}}}\) as a function of the normalised area \(\hat{A}\) of the cell (using the reference properties). The minimum value of \(\hat{G}\) corresponding to an area \({\hat{A}}_{{\text{opt}}}\) has been labelled. b The dependence of \({\hat{A}}_{{\text{opt}}}\) to the number of protein packets \({\rho }_{0}\) per unit volume available to form functional units in the cell. In both a and b, the morphological microstates are constrained to comprise only uniformly strained circular cells
At face value, stress-fibres driving cell spreading is rather counter-intuitive as stress-fibres exert contractile forces and hence one would expect them to contract the cell rather than promote spreading. To understand this apparent contradiction, recall that the number of functional units in the bound state increases with increasing strain as quantified in Eq. (5). This decreases the number of unbound stress-fibre proteins \({\hat{N}}_{{\text{u}}}\) that in turn decreases \({\chi }_{{\text{u}}}\) and therefore reduces \({\hat{F}}_{{\text{cyto}}}\). Another way to view this is to recall that the enthalpy of functional units in the bound state is lower than their corresponding enthalpy in the unbound state due to the tensile stress \({\sigma }_{{\text{max}}}\) within the stress-fibres; see Eq. (7). Chemical equilibrium dictates that all stress-fibres proteins are at equal chemical potentials. This immediately implies that the cytoskeletal free-energy decreases with increasing strain as the bound protein numbers rise with increasing strain. Thus, it is the formation of stress-fibres with tensile stresses that tends to reduce the Gibbs free-energy of the cell and drives cell spreading. Adding reagents such as cytochalasin D that inhibit the formation of stress-fibres will have the effect of diminishing the reduction in \({\hat{F}}_{{\text{cyto}}}\) with increasing strain and therefore tend to reduce cell spreading. We emphasize that cell spreading requires kinetic processes such as polymerization of meshwork actin along the cell periphery and in lamellipodia. These processes are not accounted for here but rather we argue that the overall driving force for spreading is the reduction in the Gibbs free-energy of the system and the availability of kinetic pathways is the means of achieving this reduction in the Gibbs free-energy.
3 Homeostatic mechanics for cells
The system modelled above comprised the cell and the foundation but excluded the nutrient bath that not only maintains the system at a constant temperature and pressure but also furnishes the cell with nutrients. While cells are alive they maintain a resting potential between themselves and the surrounding nutrient bath by actively regulating the concentration of various ions within the cell [30] via a very large number of complex inter-linked metabolic reactions such as (but not restricted to) ion-pumps, osmosis, diffusion and cytoskeletal reactions. Hence, from a conventional thermodynamics perspective cells are inherently in a non-equilibrium state [31]. Remarkably, the metabolic processes maintain the concentrations of all species within the cell to be very nearly constant over a sustained period of time (e.g. the interphase period of the cell cycle) and this phenomenon is known as cellular homeostasis. Shishvan et al. [22] developed a new thermodynamic framework whereby they formally defined the (dynamic) homeostatic equilibrium. We shall use this framework to analyse the distribution of states that cells assume on elastic Winkler foundations that idealise the array of micro-posts. In this section, we briefly review this homeostatic mechanics framework for cell mechanics with readers referred to [22] for details.
3.1 Brief overview of the homeostatic ensemble
The homeostatic statistical mechanics description for cells is applicable over a timescale from a few hours to a few days covering the interphase period of the cell cycle when the cell remains as a single undivided entity. Controlling only macro variables (i.e. macrostate) such as the temperature, pressure and nutrient concentrations in the nutrient bath results in inherent uncertainty (referred to here as missing information) in micro variables (i.e. microstates) of the system. This includes a level of unpredictability in homeostatic process variables, such as the spatio-temporal distribution of chemical species, that is linked to Brownian motion and the complex feedback loops in the homeostatic processes. Thus, this system not only includes the usual lack of precise information on the positions and velocities of individual molecules associated with the thermodynamic temperature, but also an uncertainty in cell shape resulting from the homeostatic processes not being precisely regulated. The consequent entropy production forms the basis of this new statistical mechanics framework motivated by the following two levels of microstates:
-
(i)
Molecular microstates Each molecular microstate has a specific configuration (position and momentum) of all the molecules within the system.
-
(ii)
Morphological microstates (Fig. 1b) Each morphological microstate is specified by the mapping (connection) of material points on the cell membrane to material points on the foundation. In broad terms, a morphological microstate specifies the shape and size of the cell.
In the homeostatic state, the system is in (dynamic) equilibrium with no net change in the internal state of the system but with a net flux of species between the system and nutrient bath (e.g. there is an overall flux of glucose into the cell while the net flow of carbon dioxide is in the opposite direction). Shishvan et al. [22] identified this (dynamic) equilibrium state by entropy maximisation. Thus, subsequently, we shall simply refer to this state as an equilibrium state to emphasise that it is a stationary macrostate of the system inferred via entropy maximisation as in a conventional equilibrium analysis. The total entropy of the system is written in terms of the conditional probability \({P}^{(i|j)}\) of the molecular microstate \((i)\) given the morphological microstate \((j)\) and the probability \({P}^{(j)}\) of morphological microstate \((j)\) as
In Eq. (19), \({I}_{{\text{M}}}^{(j)}\equiv -\sum_{i\in j}{P}^{(i|j)}\ln{P}^{(i|j)}\) and \({I}_{\Gamma }\equiv -\sum_{j}{P}^{(j)}\ln{P}^{(j)}\) are the entropies of molecular microstates in morphological microstate \((j)\) and the morphological microstates, respectively. Equilibrium then corresponds to molecular and morphological macrostates that maximise \({I}_{{\text{T}}}\) subject to the appropriate constraints. By identifying the fact that the evolution of the molecular and morphological macrostates is temporally decoupled, Shishvan et al. [22] showed that Eq. (19) can be maximised by independently maximising \({I}_{{\text{M}}}^{(j)}\) at the smaller timescales to determine the equilibrium distribution of molecular microstates (i.e. molecular macrostate) for a given morphological microstate and then maximising \({I}_{\Gamma }\) at the larger timescale to determine the equilibrium distribution of the morphological microstates (i.e. morphological macrostate).
Over the (short) timescale on the order of seconds, the only known constraint on the system is that it is maintained at a constant temperature, pressure and strain distribution. The equilibrium of a given morphological microstate \((j)\) obtained by maximising \({I}_{{\text{M}}}^{(j)}\) corresponds to molecular arrangements that minimise the Gibbs free-energy with \({G}^{(j)}\) and \({S}_{{\text{M}}}^{(j)}\) being the equilibrium values of the Gibbs free-energy and entropy of morphological microstate \((j)\), respectively. Since the connection between the cell and the foundation is fixed for a given morphological microstate, determination of \({G}^{(j)}\) is a standard boundary value problem as described in Sect. 2. Over the (long) timescale on the order of hours, the equilibrium distribution \({P}_{{\text{eq}}}^{(j)}\) is determined by maximising \({I}_{\Gamma }\) but now with the additional constraint that the cell is maintained in its homeostatic state. For the case of a cell on an elastic foundation in a constant temperature and pressure nutrient bath, the homeostatic constraint translates to the fact that the average Gibbs free-energy of the system, over all the morphological microstates it assumes, is equal to the equilibrium Gibbs free-energy \({G}_{{\text{S}}}\) of an isolated cell in suspension (free-standing cell), i.e. the homeostatic processes maintain the average biochemical state of the system equal to that of the cell in suspension. In deriving this result, Shishvan et al. [22] did not consider every individual homeostatic process but rather just used the coarse-grained outcome of the homeostatic processes, viz. over the homeostatic state, the average numbers of all species within the cell are maintained at a fixed value independent of the extracellular environment. The application of this coarse-grained constraint is the key element of the homeostatic mechanics framework with the morphological entropy \({I}_{\Gamma }\) parameterising the information lost by not modelling all the variables associated with the homeostatic processes.
The maximisation of \({I}_{\Gamma }\) while enforcing \({\sum }_{j}{P}^{(j)}{G}^{(j)}={G}_{{\text{S}}}\) gives the homeostatic equilibrium state such that
where \(Z\equiv \sum_{j}\exp\left(-\zeta {G}^{(j)}\right)\) is the partition function of the morphological microstates and the distribution parameter \(\zeta \) follows from homeostatic constraint
The collection of all possible morphological microstates that the system assumes while maintaining its homeostatic equilibrium state is referred to as the homeostatic ensemble. The homeostatic ensemble can therefore be viewed as a large collection of copies of the system, each in one of the equilibrium morphological microstates. While \({G}^{(j)}\) of each copy is not known exactly, the copies are distributed in the ensemble as per the exponential distribution \({P}_{{\text{eq}}}^{(j)}\) with the distribution parameter \(\zeta \). A crucial difference between the canonical and homeostatic ensembles is that unlike \(T\) in the canonical ensemble, \(\zeta \) is not a property of the nutrient bath but rather set by the homeostatic state that the system attains. Of course, from Eq. (20) we see that morphological microstates with lower free-energy are more probable (i.e. some sort of justification for the minimum free-energy discussion presented in Sect. 2.4) but of course the homeostatic ensemble will also contain morphological microstates with much higher free-energies, depending on the distribution parameter \(\zeta \).
The equilibrium morphological entropy \({S}_{\Gamma }=-\sum_{j}{P}_{{\text{eq}}}^{(j)}\ln{P}_{{\text{eq}}}^{(j)}\) (i.e. maximum value of \({I}_{\Gamma }\)) is related to \(\zeta \) via the conjugate relation \(\partial {S}_{\Gamma }/\partial {G}_{{\text{S}}}=\zeta \). Thus, analogous to \(1/T\) that quantifies the increase in the uncertainty of the molecular microstates (i.e. molecular entropy \({S}_{{\text{M}}}^{(j)}\)) with average enthalpy, \(\zeta \) specifies the increase in the uncertainty of the morphological microstates (i.e. morphological entropy \({S}_{\Gamma }\)) with the average Gibbs free-energy. We therefore refer to \(1/\zeta \) as the homeostatic temperature with the understanding that it quantifies the fluctuations on a timescale much slower than that characterised by \(T\).
3.2 Numerical procedure to sample the homeostatic ensemble
We employ Markov Chain Monte Carlo (MCMC) to construct a Markov chain that is representative of the homeostatic ensemble. This involves three steps: (i) a discretisation scheme to represent a morphological microstate \((j)\), (ii) calculation of \({G}^{(j)}\) for a given morphological microstate \((j)\) and (iii) constructing the Markov chain comprising these morphological microstates. Here, we briefly describe the procedure which was programmed in MATLAB with readers referred to [22] for further details.
In the general setting of a three-dimensional (3D) cell, a morphological microstate is defined by the connection of material points on the cell membrane to the surface of the foundation. In the 2D context, this reduces to specifying the connection of all material points of the cell to the foundation, i.e. a displacement field \({u}_{i}^{(j)}({X}_{i})\) is imposed on the cell with \({X}_{i}\) denoting the location of material points on the cell in the undeformed configuration and these are then displaced to \({x}_{i}^{(j)}={X}_{i}+{u}_{i}^{(j)}\) in morphological microstate \((j).\) These material points located at \({x}_{i}^{(j)}\) are then connected to material points on the foundation at the same location \({x}_{i}^{(j)}\) and this defines the morphological microstate in this 2D setting.
The cell is modelled as a continuum and thus \({u}_{i}^{(j)}\) is a continuous field. To calculate the density of the morphological microstates, we define \({u}_{i}^{(j)}\) via Non-Uniform Rational B-splines (NURBS) such that the morphological microstate is now defined by \(M\) weights \({U}_{{{L}}}^{(j)}\) (\(L=1,\dots ,M\)). In all the numerical results presented here we employ \(M=32\) with \(4\times 4\) weights governing the displacements in the \({x}_{1}\) and \({x}_{2}\) directions, respectively. The NURBS employ third order base functions for both the \({x}_{1}\) and \({x}_{2}\) directions and the knots vector included two nodes each with multiplicity three, located at the extrema of the interval. We emphasise here that this choice of representing the morphological microstates imposes restrictions on the morphological microstates that will be considered. Therefore, the choice of the discretisation used to represent \({u}_{i}^{(j)}\) needs to be chosen so as to be able to represent the microstates we wish to sample, e.g. the choice can be based on the minimum width of a filopodium one expects for the given cell type. Given \({u}_{i}^{(j)}\) we can calculate \({G}^{(j)}\) using the model described in Sect. 2.
We construct, via MCMC, a Markov chain that serves as a sample of the homeostatic ensemble. This is done using the Metropolis [32] algorithm in an iterative manner using the following procedure:
-
(i)
Assume a value of \(\zeta \) and use the undeformed cell configuration as the starting configuration and label it as morphological microstate \(j=0\) with equilibrium free-energy \({G}^{(0)}\) calculated as described above.
-
(ii)
Randomly pick two of the \(M\) weights \({U}_{{\text{L}}}^{(j)}\) and perturb them by two independent random numbers picked from a uniform distribution over the interval \([-\varDelta\,\, \varDelta ]\).
-
(iii)
Compute the new free-energy \({G}^{(j)}\) of this perturbed state and thereby the change in free-energy \(\varDelta G={G}^{(j)}-{G}^{(j-1)}\).
-
(iv)
Use the Metropolis criterion to accept this perturbed state or not, i.e.
-
(a)
if \(\varDelta G\le 0\), accept the perturbed state;
-
(b)
if \(\varDelta G>0\), compute \({P}^{{\text{acc}}}=\exp(-\zeta \varDelta G)\) and accept the perturbed state if \({P}^{{\text{acc}}}>\mathcal{R}\), where \(\mathcal{R}\) is a random number drawn from a uniform distribution over \([0 \,\,1].\)
-
(a)
-
(v)
If the perturbed state is accepted add it to the list of samples as a new morphological microstate else restore the configuration prior to step (ii) in the sample list and return to step (ii).
-
(vi)
Keep repeating this procedure until a converged distribution is obtained. Here, we typically use the criterion that the average of \({G}^{(j)}\) within the generated sample list (labelled \(\langle {G}^{(j)}\rangle \)) changes by less than 1% over 100,000 steps of the Markov chain. Typical Markov chains comprised in excess of 4 million samples.
-
(vii)
If \(\langle {G}^{(j)}\rangle \) is within \(\pm 2\%\) of \({G}_{{\text{S}}}\) we will accept this distribution else we will modify \(\zeta \) and repeat from step (i).
4 Predictions of the response of cells on a dense array of micro-posts
We present results for the response of cells on a bed of micro-posts approximated as a Winkler foundation with a normalised stiffness \(\hat{\kappa }\equiv \kappa {R}_{0}/\mu \) (see Sect. 2.3 for details of model parameters). The predictions are representative of experiments [12,13,14,15] where these is a low seeding density of cells and that there is no cell–cell interaction. Therefore, the simulations are performed for single cells. Moreover, for the range of Winkler stiffnesses \(\hat{\kappa }\) considered here we have confirmed that the cell tractions remain sufficiently small so that the two critical assumptions used within the Winkler foundation stiffness derivation (1), viz. (i) there is no post–post interaction and (ii) the post deflections are sufficiently small that linear beam theory suffices, remain valid.
The aim here is to demonstrate the application of the model in the general sense rather than aim to model a specific cell type or experiments. Hence, in the following we restrict ourselves to qualitative comparisons with existing experimental data. Predictions of the probability density functions \(p(\hat{G})\propto w(\hat{G})\exp(-\hat{\zeta }\hat{G})\) of the normalised Gibbs free-energy \(\hat{G}\) are shown in Fig. 3a for selected values of the foundation stiffness \(\hat{\kappa }\) with \(w(\hat{G})\) denoting the density of states (i.e. the fraction of total number of morphological microstates that have a normalised free-energy in the range \(\hat{G}\) to \(\hat{G}+d\hat{G}\)). Two key features emerge: (i) probability of low free-energy states decreases with decreasing foundation stiffness and (ii) the probability density functions become more peaked with decreasing foundation stiffness. The normalised homeostatic temperature \(1/\hat{\zeta }\) associated with these distributions is plotted in Fig. 3b where \(\hat{\zeta }\equiv \zeta |{G}_{{{S}}}-{\Upsilon}_{0}|\). Consistent with the more uniform distributions \(p(\hat{G})\) for the stiffer foundations, \(1/\hat{\zeta }\) increases with increasing \(\hat{\kappa }\). These results can be understood in terms of the competition between cytoskeletal and elastic energy discussed in Sect. 2.4 as follows.
Predictions of the a probability density functions \(p(\hat{G})\) of the normalised Gibbs free-energy, b the normalised homeostatic temperature \(1/\hat{\zeta }\) and c probability density functions \(p({\hat{F}}_{{\text{cyto}}})\) of the normalised cytoskeletal free-energy for selected values of the normalised foundation stiffness \(\hat{\kappa }\). In b we have indicated that cells in suspension have a zero homeostatic temperature
With increasing cell area, the concentration of bound stress-fibres increases and therefore the concentration of the unbound proteins reduces. This increases the entropy of the stress-fibre proteins and reduces their contribution to \(\hat{G}\), i.e. contribution from the cytoskeleton becomes more negative as seen from the corresponding probability distribution of \({\hat{F}}_{{\text{cyto}}}\) in Fig. 3c. On the other hand, the elastic energy of the cell increases with increasing area and this gives rise to a minimum free-energy of the cell (Fig. 2a). We shall subsequently show that cells with larger areas exert higher tractions on the foundation. However, for stiff foundations, these tractions introduce small elastic energies in the foundation and consequently the minimum system free-energy \({\hat{G}}_{{\text{min}}}\) for spread cells on stiff foundations is relatively low. By contrast, these same spread configurations introduce large elastic energies in compliant foundations with the consequence that \({\hat{G}}_{{\text{min}}}\) of the system with a soft foundation is higher than that for a stiff foundation. This implies that the system with a stiff foundation will explore free-energy configurations with a higher \(\hat{G}\) so as to compensate and maintain the average free-energy to be equal to \({G}_{{\text{S}}}\). A wider distribution \(p(\hat{G})\) with a high \(1/\hat{\zeta }\) and a mode at lower \(\hat{G}\) then ensues for stiff foundations. We emphasize that the homeostatic temperature \(1/\zeta \) is much greater than the thermodynamic temperature \(T\) for the high stiffness foundations and thus the homeostatic ensemble permits larger fluctuations than those allowed by the conventional statistical ensembles. The physical origins of these high fluctuations are the exchange of high energy species such as glucose between the cell and the nutrient bath. This exchange causes large energy fluctuations that ultimately give rise to the large observed variability in experiments as we shall proceed to show.
To illustrate the multiplicity of morphological microstates with the same free-energy, some selected configurations of the cells on the \(\hat{\kappa }=22\) and 2880 foundations are included in Fig. 4a, b, respectively. All these morphological microstates have \(\hat{G}\) values in the very close vicinity of the mode of the distribution (Fig. 3a). For each configuration, we have also included the distributions of the stress-fibre concentrations as parameterised by
Selected morphological microstates of the cells (all at the mode of the \(\hat{G}\) distribution) on foundations with normalised stiffness a \(\hat{\kappa }=22\) and b \(\hat{\kappa }=2880\). The stress-fibre concentrations as parameterised by \({\hat{N}}_{{{b}}}\) are indicated in each case. The scale bar indicates the radius \({R}_{0}\) of the undeformed cell
It is clear that even for a given fixed free-energy, the cells can attain a large diversity of cell shapes, areas and distributions of cytoskeletal proteins. Thus, even though we have only shown a very small sample of highly probable states, these results are very much in line with the diversity of observations in experiments. Moreover, in line with observations [12, 15], the images of the cells in Fig. 4a, b indicate that both the cell area and the level of actin polymerisation as parameterised by \({\hat{N}}_{{\text{b}}}\) increase with increasing foundation stiffness. In particular, cells on stiff foundations have a high concentration of polymerised stress fibres at the cell periphery and in filopodia-like structures that set the cell polarity. We now proceed to quantify the dependence of such observables on the foundation stiffness.
4.1 Foundation stiffness affects traction forces and cell spreading
We focus on two key observables that are widely reported in micro-post experiments: (i) the cell area and (ii) the traction forces. With this in mind, we define a normalised cell area as \(\hat{A}\equiv A/{A}_{0}\), where \(A\) and \({A}_{0}\) are the deformed and undeformed cell areas, respectively, while the normalised traction at location \({x}_{i}\) is
The normalised average traction then follows as
Probability density functions \(p(\hat{A})\) and \(p\left({\hat{T}}_{{\text{avg}}}\right)\) are included in Fig. 5a, b, respectively, for a range of foundation stiffnesses. Similar to \(p(\hat{G})\), \(p(\hat{A})\) and \(p\left({\hat{T}}_{{\text{avg}}}\right)\) become more peaked with decreasing foundation stiffness with the mode of the distribution simultaneously shifting to a lower \(\hat{A}\) and \({\hat{T}}_{{\text{avg}}}\). Thus, in line with experimental measurements [15] we predict that not only do the observed cell areas decrease with decreasing foundation stiffness but also the increasingly peaked distributions with decreasing \(\hat{\kappa }\) imply smaller standard errors in measurements. The overall reason for this is similar to that discussed above whereby cells on stiff foundations can spread more to lower their free-energy without introducing a large elastic penalty from the foundation and thus can sample a wider variety of morphological microstates. We note in passing that another observable typically reported in experiments is the cell aspect ratio as defined by the ratio of the major to minor axes of a best fit ellipse. For the cases investigated here the aspect ratio distribution was relatively insensitive to the foundation stiffness with the cell having a mean aspect ratio \(\approx 2.7\) in all cases.
Predictions of the probability density functions of a the normalised cell area \(\hat{A}\) and b normalised average traction \({\hat{T}}_{{\text{avg}}}\) exerted by the cell on the foundation. The probability distributions are shown for selected values of the normalised foundation stiffness \(\hat{\kappa }\)
The Markov Chain Monte Carlo calculations used in the simulations are able to determine the entire probability distribution functions (Fig. 5) by sampling in excess of few million equilibrium morphological microstates. On the other hand, experiments typically report statistics based on observations of 10–50 cell configurations [12,13,14,15] and are therefore unable to generate distributions of the type in Fig. 5. Rather, experimentalists commonly plot statistics in the form of so-called box-and-whisker diagrams. Hence to make more definitive contact with measurements, the data in Fig. 5 is re-plotted in Fig. 6a, b in the form of box-and-whisker diagrams for the distributions of \(\hat{A}\) and \({\hat{T}}_{{\text{avg}}}\), respectively. The box plots depict the median and first and third quartiles of the distributions of these observables calculated from the sample list generated by the MCMC while the whiskers show the 5th and 95th percentiles of the distributions. Clearly, the median values of \(\hat{A}\) and \({\hat{T}}_{{\text{avg}}}\) increase with increasing \(\hat{\kappa }\) with the asymptote of the infinitely stiff foundation being approximately attained for \(\hat{\kappa }>3000\). Moreover, the box-plots also clearly show that the diversity of observations increases with increasing \(\hat{\kappa }\) and this is most clearly seen in the wider spread of the whiskers with increasing \(\hat{\kappa }\).
Box-and-whisker diagrams for a the normalised cell area \(\hat{A}\) and b normalised average traction \({\hat{T}}_{{\text{avg}}}\) exerted by the cell on the foundation of normalised stiffness \(\hat{\kappa }\). The boxes show the median and the quartiles while the whiskers represent the 5th and 95th percentiles of the distributions. The normalised stiffness \(\hat{\kappa }\) is shown on the \(x\)-axis using a logarithmic scale
4.2 Correlation of traction forces and cell area
Consistent with measurements, the model predicts that the average tractions \({\hat{T}}_{{\text{avg}}}\) increase with increasing foundation stiffness. However, this rise in \({\hat{T}}_{{\text{avg}}}\) is accompanied by enhanced cell spreading (i.e. larger cell area) with increasing \(\hat{\kappa }\). Thus, it is unclear whether cells of a given area exert higher tractions on foundations with higher stiffness or if the increase seen in Fig. 6b is solely due to the increased cell area affecting \({\hat{T}}_{{\text{avg}}}\). Here we attempt to decouple these two effects.
Spatial distributions of \(\hat{T}\) for selected morphological microstates chosen from the mode of the \(\hat{G}\) distributions are shown in Fig. 7a, b for foundations with stiffness \(\hat{\kappa }=22\) and 2880, respectively (these morphological microstates are the same as in Fig. 4). Consistent with the data in Figs. 5b and 6b and a host of measurements [12,13,14,15], we see that the cells exert smaller tractions on the more complaint foundations. Moreover, as clearly seen in Fig. 7b the tractions are generally highest along the cell periphery. Of course, cells assume a large diversity of shapes and sizes even on the mode of the \(\hat{G}\) distribution (the diversity being larger for the cell on the stiffer foundation) and therefore it is not possible to decouple the effects of cell size and foundation stiffness directly from Fig. 7. We thus examine the homeostatic ensemble as sampled by the Markov chain as follows. For a given cell area, the Markov chain has a large number of morphological microstates and thus we can determine the distribution of observables for a given cell area. Here, we consider two observables, viz. the average traction \({\hat{T}}_{{\text{avg}}}\) and the nominal traction \({\hat{T}}_{{\text{T}}}\equiv {\hat{T}}_{{\text{avg}}}A/{A}_{0}\). The average and nominal tractions are equivalent to the average and total forces that are typically reported in experiments [12, 15] of cells on arrays of micro-posts.
Spatial distributions of the traction \(\hat{T}\) in selected morphological microstates of the cells (all at the mode of the \(\hat{G}\) distribution) on foundations with normalised stiffness a \(\hat{\kappa }=22\) and b \(\hat{\kappa }=2880\). The scale bar indicates the radius \({R}_{0}\) of the undeformed cell. The morphological microstates are the same as those in Fig. 4
Predictions of the variation of the three quartiles (25th percentile, median and 75th percentile) of \({\hat{T}}_{{\text{avg}}}\) and \({\hat{T}}_{{\text{T}}}\) are plotted in Fig. 8a, b, respectively, as a function of the spread cell area \(\hat{A}\) for cells on foundations with stiffness \(\hat{\kappa }=22\), 180 and 2880. For a given foundation stiffness \(\hat{\kappa }\), the dependence of \({\hat{T}}_{{\text{avg}}}\) on \(\hat{A}\) is relatively mild (Fig. 8a). As a corollary, the nominal tractions \({\hat{T}}_{{\text{T}}}\) increase nearly linearly with \(\hat{A}\) (Fig. 8b). However, it is clear from Fig. 8a that for a given cell area \(\hat{A}\), \({\hat{T}}_{{\text{avg}}}\) increases with increasing \(\hat{\kappa }\) and consequently \({\hat{T}}_{{\text{T}}}\) too is higher for cells with the same area on stiffer foundations (Fig. 8b). We thus conclude that the increased average tractions exerted by the cell on stiffer foundations are primarily due to the formation of a more concentrated stress-fibre cytoskeleton in response to stiffer foundations, but is not due to larger cell size per se. Of course the stiffer foundations also cause increased cell spreading but while this results in the cell exerting a larger total force (i.e. nominal tractions), the increased cell spreading does not significantly increase the average tractions. Finally, we note that consistent with experiments [15], even for a given cell area there is a greater diversity in the tractions exerted by cells on stiffer foundations compared to the equivalent diversity for cells on more compliant foundations (note the logarithmic axis scale for tractions in Fig. 8). Thus, the larger diversity of tractions on stiffer foundations (Figs. 5b, 6b) is not solely due to the larger diversity of cell areas that the cell assumes on stiffer foundations. This is understood by recalling that a morphological microstate is not solely characterised by a single observable such as cell area: the higher homeostatic temperature on stiffer foundations (Fig. 3b) implies that the cell assumes a great diversity of morphological microstates all with the same area and these microstates can exert a large range of tractions.
Predictions of the variation of the normalised a average traction \({\hat{T}}_{{\text{avg}}}\) and b nominal traction \({\hat{T}}_{{\text{T}}}\) as a function of the normalised cell area \(\hat{A}\) for cells on foundations with three selected values of the normalised stiffness \(\hat{\kappa }\). To illustrate the diversity of tractions that the cells of a given area can exert, we have indicated in each case the 1st quartile, median and the 3rd quartile of the distributions of the tractions
5 Concluding remarks
We have used the homeostatic mechanics framework of Shishvan et al. [22] to analyse the response of cells on a dense array of micro-posts idealised as a Winkler foundation. The framework enables the quantitative prediction of the stochastic response of cells with contractility, cell spreading and the traction forces that the cells exert on the foundation all coupled to the foundation stiffness. We show that in line with observations, the model accurately predicts that:
-
(i)
the extent of cell spreading, stress-fibre polymerisation and traction forces that the cells exert on the foundation increase with increasing foundation stiffness;
-
(ii)
the traction forces are primarily concentrated along the cell periphery; and
-
(iii)
while the total tractions increase with increasing cell area, the average tractions are reasonably independent of cell area, i.e. for a given substrate stiffness, the average tractions that are normalized by cell area do not vary strongly with cell size.
These results thus clarify the question on the source of the increased tractions with increasing foundation stiffness. In particular, they suggest that the stronger tractions that cells exert on stiffer foundations are a result of the higher levels of stress-fibre polymerisation and not directly linked to the larger levels of cell spreading that also occur on stiffer foundations.
A key feature of the model is that the predictions are statistical with cell shape, size, tractions and all other observables being outcomes of the predictions in the form of probability distributions. In fact, in line with observations, the framework predicts that the diversity of most observables such as cell area and tractions increases with increasing foundation stiffness. The homeostatic mechanics framework thus suggests that the variability in experimental observations is inherent to the homeostatic equilibrium of cells rather than being a result of in vitro experimental errors.
References
Engler AJ et al (2004) Substrate compliance versus ligand density in cell on gel responses. Biophys J 86(1):617–628
Prager-Khoutorsky M et al (2011) Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat Cell Biol 13(12):1457–1465
Lo C-M et al (2000) Cell movement is guided by the rigidity of the substrate. Biophys J 79(1):144–152
Wang N et al (2002) Micropatterning tractional forces in living cells. Cytoskeleton 52(2):97–106
Engler AJ et al (2006) Matrix elasticity directs stem cell lineage specification. Cell 126(4):677–689
Kilian KA et al (2010) Geometric cues for directing the differentiation of mesenchymal stem cells. Proc Natl Acad Sci 107(11):4872–4877
McMurray RJ et al (2011) Nanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotency. Nat Mater 10:637–644
Cramer LP, Mitchison TJ (1995) Myosin is involved in postmitotic cell spreading. J Cell Biol 131(1):179–189
Sanders LC et al (1999) Inhibition of myosin light chain kinase by p21-activated kinase. Science 283(5410):2083–2085
Legant WR et al (2010) Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat Methods 7(12):969–971
Franck C et al (2007) Three-dimensional full-field measurements of large deformations in soft materials using confocal microscopy and digital volume correlation. Exp Mech 47(3):427–438
Tan JL et al (2003) Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc Natl Acad Sci 100(4):1484–1489
Yang MT, Sniadecki NJ, Chen CS (2007) Geometric considerations of micro-to nanoscale elastomeric post arrays to study cellular traction forces. Adv Mater 19(20):3119–3123
Saez A et al (2005) Is the mechanical activity of epithelial cells controlled by deformations or forces? Biophys J 89(6):L52–L54
Han SJ et al (2012) Decoupling substrate stiffness, spread area, and micropost density: a close spatial relationship between traction forces and focal adhesions. Biophys J 103(4):640–648
Satcher RL, Dewey CF (1996) Theoretical estimates of mechanical properties of the endothelial cell cytoskeleton. Biophys J 71(1):109–118
Nelson CM et al (2005) Emergent patterns of growth controlled by multicellular form and mechanics. Proc Natl Acad Sci U S A 102(33):11594–11599
Deshpande VS, McMeeking RM, Evans AG (2006) A bio-chemo-mechanical model for cell contractility. Proc Natl Acad Sci 103(38):14015–14020
McGarry JP et al (2009) Simulation of the contractile response of cells on an array of micro-posts. Philos Trans R Soc Lond A Math Phys Eng Sci 367(1902):3477–3497
Dowling EP et al (2012) The effect of remodelling and contractility of the actin cytoskeleton on the shear resistance of single cells: a computational and experimental investigation. J R Soc Interface 9(77):3469–3479
Ronan W et al (2012) Numerical investigation of the active role of the actin cytoskeleton in the compression resistance of cells. J Mech Behav Biomed Mater 14:143–157
Shishvan SS, Vigliotti A, Deshpande VS (2018) The homeostatic ensemble for cells. Biomech Model Mechanobiol 17:1631–1662
McEvoy E et al (2018) Thermodynamic modeling of the statistics of cell spreading on ligand-coated elastic substrates. Biophys J 115:2451–2460
Buskermolen ABC et al (2019) Entropic forces drive cellular contact guidance. Biophys J 116:1994–2008
Buskermolen ABC et al (2020) Cellular contact guidance emerges from gap avoidance. Cell Rep Phys Sci 1:100055
Suresh H et al (2019) Free-energy-based framework for early forecasting of stem cell differentiation. J R Soc Interface 16:20190571
Vigliotti A et al (2016) A thermodynamically motivated model for stress-fiber reorganization. Biomech Model Mechanobiol 15(4):761–789
Ogden RW (1972) Large deformation isotropic elasticity—on the correlation of theory and experiment for incompressible rubberlike solids. Proc R Soc Lond A 326:565–584
Lucas SM, Ruff RL, Binder MD (1987) Specific tension measurements in single soleus and medial gastrocnemius muscle fibers of the cat. Exp Neurol 95(1):142–154
Keener J, Sneyd J (2009) Mathematical physiology. Springer, Berlin
Recordati G, Bellini TG (2004) A definition of internal constancy and homeostasis in the context of non-equilibrium thermodynamics. Exp Physiol 89(1):27–38
Metropolis N et al (1953) Equation of state calculations by fast computing machines. J Chem Phys 21(6):1087–1092
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare
Additional information
In honor of Professor J.N. Reddy for his 75th Birthday.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Vigliotti, A., Shishvan, S.S., McMeeking, R.M. et al. Response of cells on a dense array of micro-posts. Meccanica 56, 1635–1651 (2021). https://doi.org/10.1007/s11012-020-01208-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11012-020-01208-z