Abstract
Colorectal cancer, which is considered one of the leading causes of mortality worldwide, develops through the formation of benign polyps on the inner colon or rectum wall. Truncations in adenomatous polyposis coli (APC) gene lead to the spread of the disease in the entire colon region when combined with the guanine nucleotide exchange factor (GEF) Asef. A series of peptidomimetic agents were previously discovered as protein-protein interaction inhibitors that can target the APC-Asef interface. Structure-based virtual screening (SBVS), using a set of docking methods combined with molecular dynamics simulations, was carried out to identify new small drug-like agents. After the initial screening process, compounds with diverse chemical scaffolds and direct interaction with Arg549 and other active site residues were chosen and subjected to induce fit. The amide functional group found in the ligand hit structures showed strong interactions with Arg549, leading to observable conformational changes that allow suitable positioning within the peptide binding site. Furthermore, the pH-specific MD simulations of the top hit 838 within the APC-Asef binding site depicted significant interactions required for biochemical recognition in changing microenvironment. Predicted inhibitory constant (Ki) values and binding free energies of hits further described the significance of the amide group over the other chemical scaffolds. This combination of in silico approaches provides key insights for colorectal drug discovery programs targeting the APC-Asef interaction.

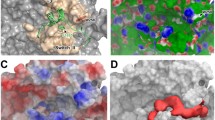
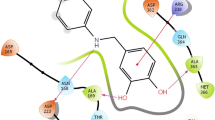
The common active site residues involved in interaction with ligands










Similar content being viewed by others
References
https://www.who.int/cancer/en/, accessed on 30th Nov, 2018
Naom S, Steven DW (2014) Colorectal polyps and polyposis syndromes. Gastroenterol Rep (Oxf) 2:1–15. https://doi.org/10.1093/gastro/got041
Jayshree M, Joe D, Sohel HQ, Satya SK, Shaw JJ, Ben C, Narendra K (2012) Prospective of colon cancer treatments and scope for combinatorial approach to enhanced cancer cell apoptosis. Crit Rev Oncol Hematol 86:232–250. https://doi.org/10.1016/j.critrevonc.2012.09.014
Jiang H, Deng R, Yang X, Shang J, Lu S, Zhao Y, Song K, Liu X, Zhang Q, Chen Y, Chen YE, Wu G, Li J, Chen G, Yu J, Zhang J (2017) Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration. Nat. Chem. Biol. 13:994–1001. https://doi.org/10.1038/nchembio.2442
Kawasaki Y, Senda T, Ishidate T, Koyama R, Morishita T, Iwayama Y, Higuchi O, Akiyama T (2000) Asef, a link between the tumor suppressor APC and G-protein signaling. Science 289:1194–1197. https://doi.org/10.1126/science.289.5482.1194
(2017) Disrupting the APC–Asef interaction suppresses cancer cell migration, Cancer Discovery, August 4, doi: https://doi.org/10.1158/2159-8290.CD-RW2017-145
Thankiah S, Wah IG, Kai PS, Kim BL, Wenyu B, Sohali A (2010) Rho GTPase Cdc42 is a direct interacting partner of adenomatous polyposis coli protein and can alter its cellular localization. Plos One 6:e16603. https://doi.org/10.1371/journal.pone.0016603
Kim SB, Zhang L, Yoon J, Lee J, Min J, Li W, Grishin NV, Wright YA, Shay JW (2018) Truncated adenomatous polyposis coli mutation induces Asef-activated Golgi fragmentation. Mol. Cell. Biol. 38:e00135–e00118. https://doi.org/10.1128/MCB.00135-18
Zhang L, Theodoropoulos PC, Eskiocak U, Moon YA, Posner B, Williams NS, Wright WE, Kim SB, Nijhawan D, De Brabander JK, Shay JW (2016) Selective targeting of mutant adenomatous polyposis coli (APC) in colorectal cancer. Sci Transl Med 8:361ra140. https://doi.org/10.1126/scitranslmed.aaf8127
Yang X, Zhong J, Zhang Q, Qian J, Song K, Ruan C, Xu J, Ding K, Zhang J (2018) Rational design and structure validation of a novel peptide inhibitor of the adenomatous-polyposis-coli (APC)-rho-guanine-nucleotide-exchange-factor-4 (Asef) interaction. J. Med. Chem 61:8017–8028. https://doi.org/10.1021/acs.jmedchem.8b01112
Lu Z, Jerry WS (2017) Multiple roles of APC and its therapeutic implications in colorectal cancer. J Natl Cancer Inst 109:djw332. https://doi.org/10.1093/jnci/djw332
Lesko AC, Goss KH, Prosperi JR (2014) Exploiting APC function as a novel cancer therapy. Curr. Drug Targets 15:90–102. https://doi.org/10.2174/1389450114666131108155418
Zhenyi Z, Leyi C, Lei G, Kui L, Liang Z, Yang L, Xiaoshan S, Yuan G, Jing Z, Ping X, Jian Z, Geng W (2011) Structural basis for the recognition of Asef by adenomatous polyposis coli. Cell Res 22:372–386. https://doi.org/10.1038/cr.2011.119
(2018) Small-molecule drug discovery suite 2018–4, Schrödinger, LLC, New York
Sterling JI (2012) ZINC 15 – Ligand discovery for everyone. J. Chem. Inf 55:2324–2337. https://doi.org/10.1021/acs.jcim.5b00559
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shaw DE, Shelley M, Perry JK, Francis P, Shenkin PS (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739–1749. https://doi.org/10.1021/jm0306430
Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL (2004) Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem 47:1750–1759. https://doi.org/10.1021/jm030644s
Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem 49:6177–6196. https://doi.org/10.1021/jm051256o
Sherman W, Day T, Jacobson MP, Friesner RA, Farid R (2006) Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem 49:534–553. https://doi.org/10.1021/jm050540c
Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY (2018) Maestro-Desmond interoperability tools. Schrödinger, New York
Kozakov D, Grove LE, Hall DR, Bohnuud T, Mottarella SE, Luo L, Xia B, Beglov D, Vajda S (2015) The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat. Protoc 10:733–755. https://doi.org/10.1038/nprot.2015.043
Kozakov D, Hall DR, Chuang GY, Cencic R, Benke R, Grove LE, Beglov D, Pelletier J, Whitty A, Vajda S (2011) Structural conservation of druggable hot spots in protein-protein interfaces. Proc. Natl. Acad. Sci. U. S. A. 108:13528–13533. https://doi.org/10.1073/pnas.1101835108
Jadav SS, Jayaprakash V, Arijit B, Sinha BN (2012) Chikungunya protease domain–high throughput virtual screening. Int Scholarly Sci Res Innovation 6:718
Jadav SS, Sinha BN, Pastorino B, De Lamballerie X, Hilgenfeld R, Jayaprakash V (2015) Identification of pyrazole derivative as an antiviral agent against chikungunya through HTVS. Lett Drug Des Discov 12:292–301. https://doi.org/10.2174/1570180811666141001005402
Xiao-Qiang Y, Zhong-Chang W, Peng-fei Q, Guigen L, Hai-Liang Z (2019) Deisgn, synthesis and biological evaluation of 2-H pyrazole derivatives containing morpholine moieties as highly potent small molecule inhibitors of APC-Asef interaction. Eur J Med Chem 177:425–447. https://doi.org/10.1016/j.ejmech.2019.05.056
Chatchakorn E, Pilkington LI, Rensburg M, White RM, Brar HK, Rees S, Paulin EK, Xu CS, Sharma N, Leung IKH, Leung E, Barker D (2020) Reynisson, Discovery of novel phosphatidylcholine-specific phospholipase C drug-like inhibitors as potential anticancer agents. Eur J Med Chem 187:111919. https://doi.org/10.1016/j.ejmech.2019.111919
Kirchner FK, Alexander EJ (1959) A new synthesis of some 2-substituted-3,4-dihydro-3-oxo-1,4,2-benzothiazine derivatives. J Am Chem Soc 81:1721–1726. https://doi.org/10.1021/ja01516a050
Ghavami M, Merino EF, Yao ZK, Elahi R, Simpson ME, Murga F, Butler JH, Casasanta MA, Krai PM, Totrov MM, Slade DJ, Carlier PR, Cassera MB (2018) Biological studies and target engagement of the 2-C-methyl-D-erythritol 4-phosphate cytidylydtransferase (SipD)-targeting antimalarial agent (1R, 3S)-MMV008138 and analogs. ACS Infec Des 4:549–559. https://doi.org/10.1021/acsinfecdis.7b00159
Lukaszuk A, Demaegdt H, Feytens D, Vanderheyden P, Vauquelin G, Tourwe D (2009) The replacemnet of His(4) in angiotensin IV by conformationally constrained residues provides highly poten and selective analogues. J Med Chem 52:5612–5618. https://doi.org/10.1021/jm900651p
Acknowledgments
SSJ is thankful for VIPER principal and CSIR-IICT Hyderabad for providing infrastructure facility.
Funding
This study is supported by the Science and Engineering Research Board (SERB) providing postdoctoral funding project (PDF/2017/001556).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
• Colorectal cancer is second leading cause of cancer death worldwide
• APC-Asef complex causes cancer cell migration in the intestine
• SBVS and MD simulations yielded 16 small molecules as APC-Asefinhibitors
• Ligand amide bonds are required for effective binding
• Arg549 can act as an amide bond detector to open the binding site
Electronic supplementary material
ESM 1
(DOCX 471 kb)
Rights and permissions
About this article
Cite this article
Jadav, S.S., Macalino, S.J.Y. & Alluri, R. Structure-based discovery of small molecule APC-Asef interaction inhibitors: In silico approaches and molecular dynamics simulations. J Mol Model 26, 207 (2020). https://doi.org/10.1007/s00894-020-04467-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-020-04467-5