Abstract
Ovarian cancer is the most lethal malignant tumor of female reproductive system. It is well-known that induction of STING-mediated type I interferons can enhance the resultant antitumor activity. However, STING pathway is usually inactivated in cancer cells at multiple levels. Here, we identified deubiquitinase USP35 is upregulated in ovarian cancer tissues. High level of USP35 was correlated with diminished CD8+ T cell infiltration and poor prognosis in ovarian cancer patients. Mechanistically, we found that silencing USP35 reinforces the activation of STING-TBK1-IRF3 pathway and promotes the expression of type I interferons. Our data further showed that USP35 can directly deubiquitinate and inactivate STING. Interestingly, activation of STING promotes its binding to USP35 in a STING phosphorylation-dependent manner. Functionally, we found that knockdown of USP35 sensitizes ovarian cancer cells to the DNA-damage chemotherapeutic drug cisplatin. Overall, our study indicates that upregulation of USP35 may be a mechanism of the restricted STING activity in cancer cells, and highlights the significance of USP35 as a potential therapeutic target for ovarian cancer.
Similar content being viewed by others
Introduction
Ovarian cancer is the leading cause of death from gynecologic cancers [1]. Nearly 300,000 new cases of ovarian cancer were diagnosed, leading to ~185,000 deaths worldwidely in 2018 [2]. The standard treatment for newly diagnosed advanced ovarian cancer is debulking surgery and platinum-based chemotherapy. Unfortunately, due to intrinsic/acquired resistant and severe adverse side-effects, a large majority of the patients are intolerable or refractory to platinum-based chemotherapy, resulting in a low overall survival rate [3, 4]. Therefore, identifying potential targets for therapeutic intervention is an urgent issue to ameliorate ovarian cancer patient outcomes.
DNA damage-based chemotherapy results in the accumulation of cytosolic DNA fragments in cancer cells [5, 6]. Cytosolic accumulated DNA may induce the production of type I interferon (IFN) via activation of Stimulator of interferon genes (STING) signaling pathway by binding to the cyclic GMP-AMP synthase (cGAS), which catalyzes the formation of the second messenger cGAMP [7, 8]. Consequently, cGAMP interacts with and activates STING, followed by the recruitment and activation of TANK-binding kinase 1 (TBK1), triggers the interferon regulator factor 3 (IRF3) and NF-κB signaling cascades and cytokines production [9, 10]. Re-activation of STING pathways may sensitize cancer cells to chemotherapy. Combination of STING agonist with carboplatin can decrease tumor burden and prolong the survival period in mice model [11]. However, the STING signaling pathway is usually inactivated in various cancers at transcriptional levels [12], whether STING is inactivated in cancer cells at post-transcriptional level remains unknown.
Ubiquitination is one of the most common protein post-translational modifications and has versatile roles in regulating cellular function [13, 14]. The conjugation of ubiquitin to a substrate protein is a multistep process mediated by an enzyme cascade, while the removal of ubiquitin moieties is catalyzed by deubiquitinating enzymes (DUBs) [15]. A number of DUBs regulate the processes associated with malignant cell phenotypes and have become potential therapeutic targets for ovarian cancer treatment. For example, USP5 has been shown to promote cell proliferation and cell cycle progression via deubiquitinating HDAC2 [16]. USP13 deubiquitinates ACLY and OGDH, drives metabolism and sensitizes tumor cells to PI3K/AKT inhibitor [17]. USP15 regulates cell viability and the stability of p53-R175H gain-of-function mutant [18]. However, the roles of DUBs in regulating STING-mediated type I interferons pathway in ovarian cancer remain poorly understood.
In the current study, we report a new function of USP35 in regulating the STING-mediated interferon signaling in ovarian cancer. We demonstrated that high expression of USP35 was correlated with diminished CD8+ T cell infiltration and worse survival rate in ovarian cancer patients. Our data collectively reveal the possible therapeutic implications of USP35 targeting as an innovative approach to ameliorate the prognosis in ovarian cancer patients.
Materials and methods
Cell lines and culture
Human HEK293T, NIH: OVCAR-3, SKOV3, Hela, murine B16F10 and CT26 cells were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The murine ovarian cancer cell line ID8 was kindly gifted from Dr Katherine F. Roby (University of Kansas Medical Center). Human OVCAR-5 was obtained from Zhongnan Hospital of Wuhan University (Wuhan, China). HEK293T, Hela, B16F10 and CT26 were maintained in DMEM medium (Gibco, USA), NIH:OVCAR-3, SKOV3 and OVCAR-5 were maintained in RPMI-1640 medium (Gibco, USA), supplemented with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, USA) and 1% penicillin-streptomycin (Invitrogen, Carlsbad, USA) at 37 °C in a humidified atmosphere incubator containing 5% CO2. The USP35 knockout (KO) primary mouse embryonic fibroblasts (MEF) were isolated from E14 embryos. The individual embryos were collected, and gonads/internal organs were removed. Then, the embryos were dispersed and trypsinized at 37 °C for 30 min. After that, the cells were isolated via centrifugation at 1000 rpm for 5 min. Then, the cells were resuspended in DMEM and were seeded on 10 cm dishes. Isolated MEFs in passage 1 were used for further experiments. Cell cultures were periodically screened for mycoplasma contamination.
Antibody, reagents and plasmids
USP35 antibody (LS-C178984) for immunohistochemistry and western blot was purchased from LifeSpan BioSciences (Seattle, USA). USP35 antibody (NBP1-28733) for immunoprecipitation was purchased from Novus Biologicals (Colorado, USA). STING (#13647), phospho-STING (Ser366, #50907), phospho-STING (Ser365, #72971), TBK1 (#3504), phospho-TBK1 (Ser172, #5483), IRF3 (#4302), phospho-IRF3 (Ser396, #4947) and normal Rabbit IgG (#2729S) were purchased from Cell Signaling Technology (Danvers, USA). CD8 antibody (ab101500) was purchased from Abcam (Cambridge, USA). Flag tag (20543-1-AP), HA tag (51064-2-AP), Myc tag (16286-1-AP), GAPDH (10494-1-AP), Tubulin (11224-1-AP) and secondary antibodies were purchased from Proteintech (Wuhan, China). Protein A/G Plus-Agarose beads (sc-2003) were obtained from Santa Cruz Biotechnology (Santa Cruz, USA). TBK1 inhibitor MRT67307 (S7948), BX-795 (S1274) and cisplatin (S1166, for in vitro experiments) were purchased from Selleck (Houston, USA). Cisplatin (for in vivo experiments) was obtained from Qilu Pharmaceutical (Shandong, China). Anti-Flag M2 agarose beads (M8823) and herring testes (HT) DNA (D6898) were purchased from Sigma-Aldrich (Darmstadt, Germany). 2′3′-cGAMP was purchased from InvivoGen (San Diego, USA). Lambda Protein Phosphatase (Lambda PP, P0753S) was purchased from New England Biolabs (Ipswich, USA). Anti-HA-tag mouse mAb (Agarose Conjugated) was purchased from Abmart (Shanghai, China). MTT (3-[4,5-dimethylthiozol-2-yl]-2,5-diphenyl tetrazolium bromide, T0793) was purchased from Sangon Biotech (Shanghai, China). Transwell® Polycarbonate Membrane (#3422) was purchased from Corning (NY, USA).
pcDNA3.1 HA, Flag, 4×Flag and Myc vectors were used to make expression plasmids. USP35 and its mutants were cloned into 5′ 4×Flag or Myc vector. STING and its mutants were cloned into 3′ Flag or HA vector as previously described [19]. cGAS was cloned into 5′ Flag vector. Ubiquitin, or ubiquitin mutants retaining a single lysine (KO) or retaining all but one lysine (KR) were cloned into pcDNA3.1 HA or Myc vector. GST-tagged STING and its mutants were cloned into pGEX-4T-2. His-tagged USP35 and its mutants were cloned into pET28a. Myc-TBK1 and IFN-β-luciferase plasmids were kindly provided by Professor Xiaojian Wang (Zhejiang University, Hangzhou, China). All point mutants and truncations were constructed by standard molecular biology techniques. All constructs were sequenced.
Bioinformatics analysis
Genetic status data available at The Cancer Genome Atlas (TCGA) database was assessed by using the cBioPortal (http://cbioportal.org) to investigate the genomic profiling of USP35 across different types of cancers [20, 21]. The Oncomine database (https://www.oncomine.org/) was used to compare the copy number of USP35 in ovarian cancer, normal ovary and blood.
Gene Expression Omnibus (GEO) database was used to compare the differential gene expression between cancer and normal cells or tissues. GSE14407 compared the expression of USP35 between 12 ovarian surface epithelial samples and 12 laser capture microdissected serous ovarian cancer epithelia samples. GSE137239 assessed the expression of USP35 in ovarian cancer cell line OVCAR-4 and OVCAR-8 when they were seeded on the 3D omentum culture compared to controls seeded on normal plastic culture dishes. GSE45553 analyzed the gene expression profiles obtained from cisplatin-sensitive OVCAR-8 and cisplatin-resistant OVCAR-8C ovarian cancer spheroids. GSE98230 compared the expression of USP35 between platinum-sensitive A2780 and platinum resistant-cells A2780cis. GSE33482 evaluated the expression data from cisplatin-sensitive A2780 and cisplatin-resistant A2780cis ovarian cancer cell lines.
Kaplan–Meier plotter (http://kmplot.com/analysis/) was used to assess the prognostic value of USP35 in patients with ovarian cancer [22]. The hazard ratio (HR) with 95% confidence intervals (CI) and log-rank p value were estimated.
The biological pathways potentially regulated by USP35 in ovarian cancer were evaluated by GSEA v3.0 software [23, 24]. Several cancer-related data sets deposited in the GSEA Molecular Signatures Database v7.0 (MSigDB) were used.
The TIMER (https://cistrome.shinyapps.io/timer/) is an online tool for systematic analysis of immune infiltrates across 32 cancer types from TCGA [25, 26]. It was used to analyze the correlation between the expression of USP35 and the abundance of immune cell infiltrates, including CD4+ T cells, CD8+ T cells, neutrophils, macrophages and dendritic cells. Additionally, the expression of USP35 in different cancer types from TCGA database was also determined by TIMER.
Human tissue microarray and immunohistochemical (IHC) analysis
The study was approved by the Institutional Ethics Committee of Tongji University Affiliated Shanghai Tenth People’s Hospital. Paired tumor and adjacent non-tumor paraffin tissue microarray for human ovarian cancer were purchased from Shanghai Zuocheng Biotech (Shanghai, China). The microarray comprised of 20 adjacent non-tumor samples, 78 serous, 17 mucinous, 10 endometrioid, 10 clear cell and 10 germ cell ovarian carcinomas samples. The diagnoses of the samples were confirmed based on the World Health Organization (WHO) classification by independent pathologists. IHC analysis was performed as previously described [27]. The staining extent was scored as: 0 (no positive cells), 1 (≤25% positive cells), 2 (26–49% positive cells), 3 (50–74% positive cells) and 4 (≥75% positive cells). The staining intensity was scored as: 0 (negative), 1 (weak), 2 (moderate) and 3 (strong). The immunoreactivity score (IRS) = extent score × intensity score, resulting in negative (0), low (1-4), medium (5–8) and high (9–12) values for each specimen. Additionally, the number of CD8+ T cells at the tumor site were counted on 5 randomly selected microscopic fields.
Generation of USP35 knockout, knockdown or overexpression cell lines
USP35 knockout ID8 cell lines were generated using lentiCRISPR methods [28]. Briefly, the oligos encoding gRNAs (USP35 sgRNA1: 5′-TTCCAGTCGCATCTACACAA-3′; USP35 sgRNA2: 5′-GGCTAAGAGTGCTGGCCTCT-3′) were constructed into the lentiCRISPRv2-puro vector. Then, the plasmids were co-transfected into HEK293T cells with packaging vectors including pSPAX2 and pMD2G. After 48 h, the culture supernatants were harvested to infect ID8 cell lines followed by two weeks of puromycin selection. To establish stable USP35 knockdown cell lines, lentiviral vector pLKO.1 expressing non-silencing shRNA control or shUSP35 were constructed. The following oligonucleotides were used: shRNA against human USP35 (5′-GGGAAGATCTGATGATGTT-3′); shRNA against mouse USP35 (5′-ACAUUGUCUUUGGAAAUGGCC-3′). To generate stable USP35 overexpression SKOV3 cell lines, human USP35 cDNA was constructed to the lentiviral expression vector pCDH-CMV-GFP-puro. Lentiviral-transduced cells were selected with puromycin (2 μg/ml) for 3 days and validated by western blotting.
Real-time quantitative PCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions, then reversed transcribed using HiScript® Q Select RT SuperMix (Vazyme Biotech, China) to synthesize cDNA samples. Quantitative real-time PCR was performed with SYBR qPCR Master Mix (Vazyme Biotech, China) and quantified by the CFX Real-Time PCR Detection System (BIO-RAD, USA). Primers used were as in Supplementary Table S1.
Cell proliferation assay
Transfected cells were collected and seeded in 96-well plates (2000 cells per well). At the end of treatment, 10 μl MTT solution (5 mg/ml) was added to each well. After 4 h of incubation at 37 °C, 200 μl dimethyl sulfoxide (DMSO) was applied to dissolve the precipitate for 30 min. Then the absorbance values were quantified at 490 nm wavelength using Tecan microplate reader (Infinite M Plex, Switzerland).
Colony-formation assay
Transfected cells were seeded in 12-well plates at an initial cell density of 500 cells per well and grown for 7–10 days with or without cisplatin. Colonies were fixed with 4% paraformaldehyde, and stained with crystal violet for 15 min at room temperature. Plates were photographed after extensive washing.
Cell migration assay
Cell migration was analyzed by using Transwell system (Corning, NY, USA) according to the manufacturer’ instruction. Cisplatin or DMSO pre-treated cells were centrifuged and resuspended in a serum-free medium. Then 200 μl cell suspension (containing 2 × 104 cells) were added to the upper Transwell chambers with or without cisplatin. The lower chamber contained 800 μl of medium with 15% FBS. After incubation at 37 °C for 24 h, the migrated cells on the lower surface of the chamber were fixed with 4% paraformaldehyde and stained with crystal violet.
Immunoprecipitation (IP) and western blotting
Transfected cells were collected and lysed in lysis buffer (150 mM NaCl, 0.5% NP-40, 1 mM EDTA, 10% glycerophosphate, 50 mM Tris-Cl, pH 7.4 and a cocktail of proteinase inhibitors). After 30 min, the cell lysates were isolated via centrifugation at 12,000 rpm 4 °C for 15 min. For IP, cleared cell lysates were incubated with M2 beads or HA-conjugated beads at 4 °C for 90–120 min. To detect endogenous protein interactions, cell lysates were incubated with 15 μl of protein A/G agarose beads and indicated antibody at 4 °C overnight. The beads were boiled after extensive washing, resolved via SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed via immunoblotting. The immunoblots were detected using the electrochemiluminescence (ECL) imaging system (Tanon, Shanghai, China) or the Odyssey system (LI-COR Biosciences, Lincoln, USA).
Deubiquitination assay
For the in vivo ubiquitination assay, HEK293T cells were transiently co-transfected with indicated plasmids. After 24 h, cells were lysed with 100 μl lysis buffer (2% SDS, 150 mM NaCl and 10 mM Tris-HCl, pH 8.0), boiled for 20 min. 900 μl dilution buffer (150 mM NaCl, 1% Triton, 2 mM EDTA and 10 mM Tris-HCl, pH 8.0) was added. The samples were incubated with M2 or HA beads at 4 °C for 90–120 min with rotation. Then the beads were boiled after extensive washing with washing buffer (1 M NaCl, 1% NP-40, 1 mM EDTA and 10 mM Tris-HCl, pH 8.0), and resolved via SDS-PAGE gel for immunoblotting analysis.
To detect the in vitro deubiquitination of STING, His-USP35 WT and CA proteins were purified from E. coli. HEK293T cells were co-transfected with indicated plasmids. After 24 h transfection, cell lysates were boiled and immunoprecipitated using M2 beads. Then the reaction system was added with His-USP35 WT or CA protein and incubated at 37 °C for 4 h. The ubiquitinated STING proteins were detected by immunoblotting analysis. All ubiquitination tests were using denature-immunoprecipitation assay.
GST pull-down assay
The His-USP35 protein was purified from E. coli and incubated with purified GST or GST-STING protein. The GST proteins were purified using glutathione sepharose 4B. The beads were washed extensively, and the bound USP35 was detected by immunoblotting analysis.
Phosphatase treatment
Flag-USP35 and STING-HA plasmids were co-expressed in HEK293T cells. After 24 h transfection, the cells were treated with cGAMP at indicated time. Cells were lysed and immunoprecipitated using M2 beads. Then the protein samples were combined with 10× NEBuffer, 10 mM MnCl2 and Lambda Protein Phosphatase (New England Biolabs), and incubated at 30 °C for 60 min. The reaction mixtures were boiled and detected by immunoblotting with indicated antibodies.
Luciferase reporter analysis
HEK293T cells were co-transfected with IFN-β firefly luciferase reporter plasmid, Renilla luciferase plasmid, STING and USP35 expression plasmids by polyethylenimine (PEI) according to the manufacturer’s instruction. After 24 h, cells were lysed and centrifugated, and luciferase activity was measured using the dual-luciferase reporter assay system (Promega, Madison, USA). Relative IFN-β activity was calculated as firefly luminescence relative to Renilla.
Tumor xenografts
The animal experiment was administered according to the guidelines of Institution Animal Care and Use Committee and the protocol was approved by Shanghai Tenth People’s Hospital affiliated to Tongji University. 5 × 106 shNC or shUSP35 ID8 cells in 200 μl of PBS were injected into the abdominal cavities of 6–8 week old female C57BL/6 mice (Shanghai SLAC Laboratory Animal Co.,Ltd, China). The four groups (n = 24) were randomized and categorized based on the treatment regime as: shNC + PBS (n = 6), shUSP35 + PBS (n = 6), shNC + cisplatin (n = 6) and shUSP35 + cisplatin (n = 6). 3-weeks post ID8 cells implantation, cisplatin (2 mg/kg) or PBS was administered intraperitoneally on days 21, 28, 35 and 42. Tumor progression was monitored by body weight. The mice were sacrificed when the control mice reached endpoint (abdominal diameter ≥35 mm). The disseminated tumor weight and ascites volume were evaluated, and tumor tissues were collected for further analyses.
Statistical analysis
GraphPad Prism software (Version 6.01, GraphPad Software, Inc., USA) was used for data analysis. All grouped data are presented as mean ± SD and statistical significance was determined by using Student’s t tests. Survival rates were assessed using Kaplan–Meier curves and the log-rank test. All the experiments were repeated independently at least three times. P value < 0.05 was considered statistically significant.
Results
Amplification of USP35 in ovarian cancer
To explore the role of DUBs in ovarian cancer, we investigated the genomic and expression alterations of DUBs family. By analyzing the TCGA genomic data, we found that USP35 is one of the top-ranked amplified deubiquitinase genes in ovarian cancer (Fig. 1a, Supplementary Fig. S1A), and the amplification is the most common genetic event of USP35 in ovarian cancer (Fig. 1b). Oncomine database revealed that the copy number of USP35 is highly elevated in ovarian cancer than in normal ovary or blood samples (Supplementary Fig. S1B). Consistently, the mRNA levels of USP35 in ovarian cancer epithelia samples are significantly higher than in ovarian surface epithelia samples (GSE14407, Supplementary Fig. S1C). Moreover, USP35 mRNA is also upregulated once ovarian cancer cell line OVCAR-4 and OVCAR-8 are seeded on the three-dimensional omentum culture model mimicking early metastasis (GSE137239, Supplementary Fig. S1D). Except in ovarian cancer, USP35 expression is also upregulated in several other cancer types, such as breast invasive carcinoma, colon adenocarcinoma, head and neck cancer, lung adenocarcinoma and liver hepatocellular carcinoma compared with corresponding normal adjacent tissues (Supplementary Fig. S1E).
a Genomic profiling of USP35 across human cancers determined by cBioPortal (http://www.cbioportal.org/) of TCGA databases. b Scatterplots of USP35 mRNA expression versus copy number in ovarian cancer from cBioPortal. c Representative pictures of USP35 IHC staining in ovarian cancer and adjacent non-tumor tissues (ANT). Scale bar = 500 μm (upper) and 50 μm (lower). d IHC score of USP35 in different subtypes of ovarian cancer (serous, n = 78; mucinous, n = 17; endometrioid, n = 10; clear cell, n = 10; germ cell, n = 10) and ANT (n = 20). e The percentage of USP35 staining in different subtypes of ovarian cancer. f Representative pictures of USP35 IHC staining in different grades of ovarian cancer. Scale bar = 500 μm (upper) and 50 μm (lower). g IHC score of USP35 in different grades of ovarian cancer. h Overall survival (OS) and progression-free survival (PFS) curves in USP35 high and low expression ovarian cancer cases from Kaplan–Meier plotter database (http://kmplot.com/analysis/). i Kaplan–Meier OS and PFS curves in USP35 high and low expression ovarian cancer cases from GSE9891. j Kaplan–Meier OS and PFS curves in USP35 high and low expression ovarian cancer patients with platin-chemotherapy from GSE9891. Error bar ± SD, *p < 0.05.
To further confirm that USP35 is upregulated in ovarian cancer, we examined the expression of USP35 in ovarian cancer tissues by IHC analysis. As shown in Fig. 1c, d, USP35 protein levels were significantly higher in ovarian cancer tissues compared with adjacent normal tissues (ANT) (mean IHC socre: 4.35 [ANT] vs. 10.22 [serous], 9.06 [mucinous], 10.00 [endometrioid] and 8.60 [clear cell], p < 0.05). In contrast, no difference was detected between ovarian germ cell tumor tissues and ANT (mean IHC socre: 4.35 [ANT] vs. 5.00 [germ cell], p = 0.62) (Fig. 1c, d). Serous, mucinous and endometrioid carcinomas exhibited strong positive staining, followed by clear cell and germ cell carcinomas (Fig. 1e). Moreover, the protein level of USP35 was also positively correlated with tumor grade (Fig. 1f, g).
We also evaluated the predictive value of USP35 for prognosis in ovarian cancer. Kaplan–Meier survival analysis revealed that high expression of USP35 was correlated with poor overall survival (OS) (p = 0.0358, HR = 1.29, 95% CI = 1.02–1.65) and progression-free survival (PFS) (p = 4.3e-5, HR = 1.60, 95% CI = 1.27–2.00) (Fig. 1h). GSE9891 showed that high expression of USP35 predicted worse OS (p = 0.0015, HR = 2.25, 95% CI = 1.35–3.75) and PFS (p = 0.0063, HR = 1.61, 95% CI = 1.14–2.28) in ovarian cancer patients (Fig. 1i). Besides that, USP35 upregulation was associated with shorter OS (p = 0.0095, HR = 1.94, 95% CI = 1.17–3.24) and PFS (p = 0.0433, HR = 1.44, 95% CI = 1.01–2.05) in patients with platin-chemotherapy (GSE9891, Fig. 1j). Together, these data suggest that USP35 may be a potential positive regulator of ovarian cancer development.
Overexpression of USP35 is correlated with reduced immune cell infiltration
To explore the potential pathological roles of USP35 in ovarian cancer, we analyzed the correlation between immune cell infiltration and USP35 expression. Interestingly, our data from immunohistochemical stains showed that a preferential decrease in intratumoral CD8+ T cell infiltration in ovarian cancer specimens with high expression of USP35 (Fig. 2a, b). We confirmed the infiltration of CD8+ T cell in ovarian cancer via TIMER platform (Fig. 2c). Moreover, overexpression of USP35 was also negatively correlated with infiltration of macrophage (Fig. 2d), neutrophil (Fig. 2e) and dendritic cell (Fig. 2f), but not CD4+ T cell and B cell (Supplementary Fig. S2). Importantly, similar correlations were also observed across several other types of cancers (Fig. 2c–f). These data suggest that USP35 may contribute to immune regulation in ovarian cancer.
a, b Representative images and analysis from high or low IHC scores of USP35 in ovarian cancer tissues. Red arrowheads indicate positive CD8+ T cells. Scale bar = 500 μm (left) and 50 μm (right). Correlation between the expression of USP35 and infiltrating level of CD8+ T cells (c), macrophage (d), neutrophil (e) and dendritic cell (f) across different types of cancers based on TIMER platform (https://cistrome.shinyapps.io/timer/). ACC adrenocortical carcinoma, BLCA bladder urothelial carcinoma, BRCA breast invasive carcinoma, CESC cervical squamous cell carcinoma and endocervical adenocarcinoma, COAD colon adenocarcinoma, DLBC lymphoid neoplasm diffuse Large B-cell Lymphoma, ESCA esophageal carcinoma, GBM glioblastoma multiforme, HNSC head and neck squamous cell carcinoma, KICH kidney chromophobe, KIRC kidney renal clear cell carcinoma, KIRP kidney renal papillary cell carcinoma, LIHC liver hepatocellular carcinoma, LUAD lung adenocarcinoma, LUSC lung squamous cell carcinoma, OV ovarian serous cystadenocarcinoma, PAAD pancreatic adenocarcinoma, PRAD prostate adenocarcinoma, READ rectum adenocarcinoma, STAD stomach adenocarcinoma, THCA thyroid carcinoma, UCEC uterine corpus endometrial carcinoma, UVM uveal melanoma.
Silencing of USP35 potentiates STING-mediated interferon signaling in ovarian cancer cells
To explore the potential mechanism of negative correlation between the USP35 expression and T cells infiltration, we analyzed the USP35-related signaling pathways using GSEA analysis of TCGA data for ovarian cancer. Our data showed that RESPONSE TO INTERFERON BETA DN (NES = −1.65, p < 0.05) and GRANDVAUX IRF3 TARGETS DN (NES = −1.35, p < 0.01) were significantly enriched in patients with lower levels of USP35 expression (Fig. 3a, b). This is consistent with the function of interferon signaling in cancer cells in recruitment of immune cells and antitumor efficacy [29, 30]. These data suggest that upregulation of USP35 may correlate with interferon pathway.
a, b GSEA analysis showed that the gene sets of NATSUME RESPONSE TO INTERFERON BETA DN and GRANDVAUX IRF3 TARGETS DN were enriched in the USP35-low group. qPCR analysis of Ifn-β, Cxcl10 or Isg15 mRNA in USP35+/+ and USP35−/− ID8 cells followed by treatment with HT-DNA for 4 h (c) or cGAMP for 2 h (d). qPCR analysis of IFN-β, CXCL10 or ISG15 mRNA in SKOV3 (e) or OVCAR-3 (f) cells stably transfected with shNC or shUSP35 followed by treatment with cGAMP for 2 h. g qPCR analysis of Ifn-β, Cxcl10 or Isg15 mRNA in USP35+/+ and USP35−/− MEF cells followed by treatment with cGAMP for 2 h. Data are representative of three independent experiments. Error bar ± SD, *p < 0.05. NES normalized enrichment score, FDR false discovery rate.
To examine whether USP35 is indeed involved in the regulation of interferon pathway, we transfected ID8 cells with USP35-specific Cas9/sgRNA or shRNA and the production of type I interferon was examined. Our data showed that both HT-DNA and cGAMP-triggered expression of Ifn-β, as well as Cxcl10 and Isg15, were significantly enhanced in USP35 knockout or knockdown ID8 cells (Fig. 3c, d, Supplementary Fig. S3A, B). Similar results were obtained in human ovarian cancer SKOV3, OVCAR-3, OVCAR-5 cells, human cervical cancer Hela cells, mouse melanoma B16F10 cells, and mouse colorectal carcinoma CT26 cells once USP35 was knockdown by shRNA (Fig. 3e, f, Supplementary Fig. S3C–F). To further confirm this result, we obtained the USP35 knockout MEFs and examined its role in interferon signaling. Our data showed that the expression of Ifn-β, Cxcl10 or Isg15 was significantly increased in USP35−/− MEFs compared to wild-type MEFs in response to cGAMP (Fig. 3g). Furthermore, overexpression of USP35 abolished cGAMP-triggered induction of IFN-β, CXCL10 and ISG15 in SKOV3 cells (Supplementary Fig. S4A). Collectively, these data suggest that USP35 is a negative regulator of interferon signaling in cancer cells.
USP35 interacts with and deubiquitinates STING
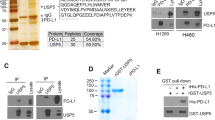
It is well-known that activation of cGAS-STING-interferon signaling is correlated with infiltration of antitumor T cells in cancer tissues [31,32,33]. Moreover, STING is the major identified DNA and cGAMP sensor in cells. We therefore examined whether USP35 has any role in STING-mediated interferon signaling by using luciferase reporter and qPCR assays. Our data showed that USP35 inhibited the activation of IFN-β reporter and mRNA expression of IFN-β mediated by STING (Fig. 4a, b). We also found that wild-type (WT), but not enzymatic inactive mutant (C450A) USP35, blocked STING-induced IFN-β reporter activation and mRNA expression (Fig. 4c, d).
a–d Indicated plasmids were transfected in HEK293T for 24 h, IFN-β luciferase activity and IFN-β mRNA expression were measured. STING-HA and Myc-USP35 protein levels were shown. e STING-Flag was transfected into HEK293T cells in combination with Myc-USP35WT or Myc-USP35C450A and HA-ubiquitin for 24 h. Cell lysates were immunoprecipitated with M2 beads. A denaturation assay was employed to test ubiquitination. f Hydrolysis of ubiquitin chains conjugated to STING by USP35 was analyzed using an in vitro deubiquitination assay. g Denature-IP (with anti-HA) and immunoblot analysis (with anti-Flag, anti-HA or anti-Myc) of HEK293T cells transfected with indicated plasmids for 24 h. h Immunoprecipitation (IP, with anti-Flag) and immunoblot analysis (with anti-Flag, anti-Myc) of HEK293T cells transfected with plasmids encoding Flag-tagged STING or cGAS and Myc-USP35 for 24 h. i Endogenous USP35 was immunoprecipitated with an antibody against USP35 from ID8 cells, and the associated STING was detected by an anti-STING antibody. j Schematic showing domain structures of STING and USP35 as indicated in the text. IP (with anti-Flag) and immunoblot analysis (with anti-Myc, anti-Flag or anti-HA) of HEK293T cells transfected with plasmids encoding Myc-USP35 and Flag-tagged STING or STING truncates (k) or transfected with plasmids encoding STING-HA and 4Flag-tagged USP35 or USP35 truncates (l). Error bar ± SD, *p < 0.05. ER endoplasmic reticulum, NTD N-terminal repeat domain, CTD C-terminal repeat domain.
It is known that ubiquitination plays an essential role in STING activation. We therefore examined whether USP35 serves as a deubiquitinase of STING. As shown in Fig. 4e, USP35WT, but not USP35C450A, catalyzed deubiquitination of STING. The in vitro deubiquitination assay also confirmed that STING could be deubiquitinated by USP35 directly (Fig. 4f). To explore the type of polyubiquitin linkage on STING which was modified by USP35, we co-transfected STING with WT-ubiquitin, or ubiquitin KO or KR mutants. Our data showed that USP35 could remove K6-, K11-, K27-, K29- or K63-linked polyubiquitin chains from STING (Fig. 4g, Supplementary Fig. S5). Together, these results indicate that USP35 can function as a deubiquitinase for STING.
Then we examined whether USP35 is a direct binding partner of STING. We performed a co-IP assay and revealed that STING, but not cGAS, bound to USP35 (Fig. 4h). The association between endogenous USP35 and STING proteins was also validated by immunoprecipitation (Fig. 4i). To map the domains that mediate the interaction between USP35 and STING, series of truncation mutants were expressed in HEK293T cells. Our co-IP assay showed that only the cytoplasmic region (C-terminal domain, CTD, amino acids 137–379) of STING bound to USP35 (Fig. 4j, k). The C-terminal USP catalytic domains (including amino acids 433–603, 433–753, 754–1018 and 433–1018), but not the N-terminal HEAT repeats region (amino acids 1-432) of USP35 were responsible for its interaction with STING (Fig. 4j, l). Taken together, our results indicate that USP35 functions as a deubiquitinase and can directly target STING for deubiquitination.
USP35 regulates STING-TBK1-IRF3 pathway
Next, we examined whether USP35 regulates STING-mediated signaling. STING activation induces the recruitment of TBK1 and triggers IRF3 signaling cascades [9, 10]. Our data showed that depletion of USP35 potentiated HT-DNA and cGAMP-induced phosphorylation of STING, TBK1 and IRF3 in ID8 cells (Fig. 5a, b, Supplementary Fig. S6A, B). Similar enhanced phosphorylation of STING, TBK1 and IRF3 were obtained in USP35 knockdown SKOV3, OVCAR-3 and OVCAR-5 cells in response to cGAMP stimulation (Fig. 5c, d, Supplementary Fig. S6C). Moreover, the phosphorylation of STING, TBK1 and IRF3 were also increased in USP35−/− MEFs (Fig. 5e), USP35 depleted B16F10, CT26 and Hela cells (Supplementary Fig. S6D–F) after treatment with cGAMP. In addition, USP35 overexpression abolished cGAMP-induced phosphorylation of STING, TBK1 and IRF3 in SKOV3 cells (Supplementary Fig. S4B). Previous studies have revealed that K27- or K63- linked STING polyubiquitination is required for the recruitment of TBK1 [34, 35], we therefore investigated whether USP35 affects the STING-TBK1 interaction. Our data showed that knockdown of USP35 in ID8 cells enhanced the endogenous binding of STING to TBK1 upon HT-DNA transfection (Fig. 5f). In contrast, overexpression of USP35WT, but not USP35C450A, inhibited STING-TBK1 association in HEK293T cells (Fig. 5g). Moreover, ectopic expression of USP35WT, but not USP35C450A, reduced phosphorylation and multimerization of STING (Fig. 5h, i). These data suggest that enzymatic activity of USP35 is essential for mediating the STING-TBK1-IRF3 pathway.
Immunoblot analysis of phosphorylation of STING, TBK1 and IRF3, total STING, TBK1 and IRF3 in USP35+/+ and USP35−/− ID8 cells followed by treatment with HT-DNA for 0–4 h (a) or cGAMP for 0–2 h (b). Immunoblot analysis of phosphorylation of STING, TBK1 and IRF3, total STING, TBK1 and IRF3 in SKOV3 (c) or OVCAR-3 (d) cells stably transfected with shNC or shUSP35 followed by treatment with cGAMP for 0–2 h. e Immunoblot analysis of phosphorylation of STING, TBK1 and IRF3, total STING, TBK1 and IRF3 in USP35+/+ and USP35−/− MEF cells followed by treatment with cGAMP for 0–2 h. f shNC or shUSP35 ID8 cells were transfected with HT-DNA for 0–4 h. Cell lysates were immunoprecipitated with an antibody against STING. All proteins were immunoblotted with the indicated antibodies. g Flag-TBK1 was co-transfected with STING-HA and Myc-USP35WT or Myc-USP35C450A into HEK293T cells for 24 h. Cell lysates were immunoprecipitated with HA beads. All proteins were immunoblotted with the indicated antibodies. h STING-Flag was co-transfected with Myc-USP35WT or Myc-USP35C450A into HEK293T cells for 24 h. Phosphorylation of STING and TBK1, total STING and TBK1 were analysis by western blotting. i STING-HA were co-transfected with STING-Flag and Myc-USP35WT or Myc-USP35C450A into HEK293T for 24 h. STING-Flag was immunoprecipitated using M2 beads. The bound STING-HA was analyzed by western blotting.
Phosphorylation of STING at Ser366 is essential for USP35-STING association
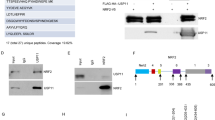
Next, we explored whether the association between USP35 and STING was dependent on the activation of STING pathway. To this end, we activated STING by HT-DNA transfection or cGAMP stimulation and examined the association between USP35 and STING. We observed their biochemical interaction were enhanced once STING was activated (Fig. 6a, b, Supplementary Fig. S7A). We then examined the mechanism by which the enhanced interaction induced by STING activation. Because activation of STING pathway is accompanied by the phosphorylation of STING [36], we examined whether the interaction between USP35 and STING is dependent on the phosphorylation. Our data showed that lambda protein phosphatase treatment decreased the interaction between USP35 and STING enhanced by cGAMP (Fig. 6c), indicating that the association between these two proteins was phosphorylation-dependent. Ser366 is the major phosphorylation site of STING, which is catalyzed by TBK1 [37]. We therefore examined whether the Ser366 phosphorylation of STING is involved in its interaction with USP35. Our data showed that compared to STINGWT, STINGS366A mutant exhibited a reduced affinity for USP35 (Fig. 6d, e). Moreover, exogenous co-expression of TBK1 increased their interaction (Fig. 6f), whereas treatment with TBK1 inhibitor MRT67307 or BX795 reduced their interaction induced by cGAMP (Fig. 6g, Supplementary Fig. S7B). Moreover, USP35 exhibited reduced activity to deubiquitinate STINGS366A mutant than wild-type STING (Fig. 6h). Taken together, these data suggest that activation of STING promoted its binding to USP35 in a phosphorylation-dependent manner, which in turn led to STING deubiquitination and reduced the production of type I interferon.
a ID8 cells were transfected with HT-DNA for 0–2 h. Endogenous USP35 was immunoprecipitated with an antibody against USP35, and the associated STING was detected by an anti-STING antibody. b STING-HA was transfected into HEK293T cells in combination with 4Flag-USP35 for 24 h. After treatment with cGAMP for 0–2 h, cell lysates were immunoprecipitated with M2 beads. All proteins were immunoblotted with the indicated antibodies. c Indicated plasmids were co-transfected into HEK293T cells. After treatment with cGAMP for 0–2 h, cell lysates were immunoprecipitated with M2 beads and incubated with Lambda Protein Phosphatase. All proteins were immunoblotted with the indicated antibodies. d Immunoprecipitation (with anti-Flag) and immunoblot analysis (with anti-Flag or anti-HA) of HEK293T cells transfected with plasmids encoding Flag-USP35 and HA-tagged STING WT, S366D or S366A mutant. e The interaction between USP35 and STING WT, S366D or S366A mutant (amino acids 137–379) was assessed using a GST pull-down assay. All proteins were detected using the indicated antibodies. f STING-HA was transfected into HEK293T cells in combination with 4Flag-USP35 and Myc-TBK1. After 24 h, cell lysates were immunoprecipitated with M2 beads. All proteins were immunoblotted with the indicated antibodies. g Indicated plasmids were co-transfected into HEK293T cells. Cells were pre-treated with or without TBK1 inhibitor MRT67307 (2 μM) for 2 h and then administrated with cGAMP. The cell lysates were immunoprecipitated with M2 beads. All proteins were immunoblotted with the indicated antibodies. h Denature-IP (with anti-HA) and immunoblot analysis (with anti-Flag, anti-HA or anti-Myc) of HEK293T cells transfected with indicated plasmids for 24 h. All proteins were immunoblotted with the indicated antibodies.
USP35 regulates cisplatin-induced STING-mediated interferon in ovarian cancer
To gain insight into the significance of USP35-modulated STING-interferon pathway for ovarian cancer therapy, we performed GSEA with TCGA database to explore the correlation between USP35 and gene sets that are associated with chemotherapy effectiveness in ovarian cancer. Interestingly, USP35 was negatively correlated with the gene signature of LI CISPLATIN RESISTANCE DN (NES = −1.41, p < 0.01) (Supplementary Fig. S8A). Moreover, USP35 was also significantly elevated in cisplatin-resistance ovarian cancer cells (Supplementary Fig. S8B–D). In view of cisplatin is a DNA-damage chemotherapeutic drug, we speculated whether USP35 could regulate cisplatin-induced STING-mediated interferon in ovarian cancer. To this end, we performed xenograft studies using shNC and shUSP35-ID8 cells injected intraperitoneally into female C57BL/6 mice. After 3 weeks, the mice were treated with cisplatin (2 mg/kg weekly) or PBS intraperitoneally. On day 56, the mice were killed for further analysis (Fig. 7a). As peritoneal metastasis and malignant ascites are the hallmarks of advanced ovarian cancer [38], we examined peritoneal dissemination and ascites production in the mouse model. Our data showed that the body weight and ascites volume of mice were lower in the shUSP35 group (mean body weight: 27.72 g; mean ascites volume: 9.00 ml) than the shNC group (mean body weight: 29.33 g; mean ascites volume: 11.50 ml) at the end point of the experiment (Fig. 7b–d). Reduced ascites accumulation was observed in the cisplatin-treated shUSP35 group (mean body weight: 18.27 g; mean ascites volume: 1.92 ml) compared to the cisplatin-treated shNC group (mean body weight: 20.38 g; mean ascites volume: 3.67 ml) (Fig. 7b–d). Meanwhile, silencing of USP35 resulted in profound decrease in tumor dissemination and tumor weight in the cisplatin-treated shUSP35 group (Fig. 7e–g). Moreover, the expressions of Ifn-β and Cxcl10, but not other cisplatin-sensitivity related genes, were enhanced in tumors derived from shUSP35 cells compared to shNC cells treated with cisplatin (Fig. 7h, Supplementary Fig. S9). Consistently, knockdown of USP35 also sensitized ID8, OVCAR-5 and Hela cells to cisplatin and promoted the expression of IFN-β and CXCL10 in response to cisplatin in vitro (Supplementary Fig. S10A–F). Thus, our data confirmed that silencing USP35 potentiates cisplatin-induced expression of interferons in ovarian cancer.
a Summary of experimental design to study the effect of USP35 knockdown in combination with cisplatin in ovarian cancer is shown. b Body weights of mice were measured on indicated days. c, d Ascites volumes (ml) were measured when the shNC group mice reached endpoint (abdominal diameter of ≥35 mm; 6 mice per group). e–g Tumor formation in mice transplanted with ID8 cells expressing control shRNA or USP35 shRNA, combined with or without treatment of cisplatin (2 mg/kg). Representative images show tumor dissemination and tumor weights. h qPCR analysis of Ifn-β and Cxcl10 in tumors derived from ID8 cells. i Model showing that USP35 negatively regulates cisplatin sensitivity in ovarian cancer through regulating STING-mediated interferon signaling. Error bar ± SD, *p < 0.05.
Discussion
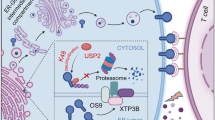
Although activation of STING-associated type I interferons signaling has been proven to enhance the resultant antitumor activity, inactivation of STING pathway is usually found in various cancers. In current study, we demonstrated that activation of STING pathway induced the recruitment of a cancer overexpressed deubiquitinase USP35 to STING, which leads to deubiquitination of STING and suppression of type I interferons signaling. The data presented here provided the mechanism and potential therapeutic strategy in which silencing USP35 potentiates cytosolic DNA and cGAMP-induced expressions of type I interferon gene and down-stream chemokines through STING-TBK1-IRF3 pathway, and also reinforces cisplatin-induced expression of interferons in ovarian cancer. Overall, this study reveals a novel regulatory function of USP35 in STING-mediated interferon pathway in ovarian cancer (Fig. 7i).
It has been well documented that DNA damage arising from exogenous stresses (such as genotoxic anticancer drugs) activates the cGAS-STING-TBK1 signaling, resulting in upregulation of cytotoxic interferons [6, 10, 39]. Accumulating evidences suggest that targeting STING pathway increases the efficacy of antitumor response in ovarian cancer. Ghaffari et al. reported that treatment with STING agonist reduced ascites formation and tumor burden, and its combination with carboplatin prolonged the survival time of mice [11]. Furthermore, PARP inhibition promotes accumulation of cytosolic DNA fragments and generates local/systemic antitumor responses through STING-dependent pathway in mice bearing tumors derived from BRCA1-deficient ovarian cancer cells [6, 40]. However, cancer cells often exhibit deficiency of STING signaling and suppression of interferon production to evade immune detection [41, 42]. In this study, we found that activation of STING, which induces the phosphorylation of STING at Ser366, promoted its binding to USP35. As USP35 is upregulated in ovarian cancer, our study provides a mechanism by which STING is inactivated in cancers, especially upon treatment with DNA-damage chemotherapeutic drug cisplatin.
The post-translational modifications, especially ubiquitination, are critical for the interferon-stimulating activity of STING to regulate immune responses [43]. But how the STING is regulated at post-translational modifications in cancers remains largely unknown. In this study, we identified USP35 as a negative regulator of STING-associated type I interferons signaling at post-translational modification in ovarian cancer cells. We observed that silencing of USP35 potentiates cytosolic DNA and cGAMP-induced expressions of type I interferon gene IFN-β and down-stream chemokines such as CXCL10 and ISG15 through STING-dependent pathway in ovarian cancer cells. Our study is consistent with a previous systematic screening research for the functions of DUBs in regulating antiviral responses, which revealed that USP35 can modulate type I interferons and regulate the replication of viruses [44]. We also noticed that fold difference of IFN-β, CXCL10 and ISG15 mRNA upon USP35 knockout or knockdown in ovarian cancer cells is mildly significant, suggesting that USP35 partially suppresses STING-mediated signaling. Because several deubiquitinases, such as USP13, USP20, USP21, USP49 and CYLD, have been reported as a deubiquitinase for STING [19, 45,46,47,48], it will be interesting to study whether STING is also dynamically regulated by these DUBs in cancers.
Over the past decades, STING-mediated type I interferons production has been demonstrated to possess antitumor effect by inhibiting cancer cell proliferation, regulating chemosensitivity and participating in cancer immunoediting [49,50,51,52]. Importantly, intraperitoneally administration of type I interferons suppressed the formation of malignant ascites in the model of ovarian cancer xenograft mice, even in ovarian cancer patients [53,54,55]. Previous studies indicate that cytosolic double-stranded DNA (dsDNA) triggers interferons response and antitumor immune signaling [56, 57]. As a DNA-damage-inducing drug, cisplatin is commonly used for treatment of ovarian cancer, and it has been reported to promote immune-mediated tumor elimination through STING-associated interferon signaling [58, 59]. Moreover, the combination of interferons and cisplatin exhibited synergistic inhibitory effects on the tumor growth [50, 60, 61]. In our mice ovarian cancer model, knockdown of USP35 in ID8 cells with a combination treatment of cisplatin showed significant suppression of peritoneal dissemination and ascites formation. We also observed the increased IFN-β and CXCL10 expressions in tumors derived from cisplatin-treated shUSP35 cells compared to shNC cells. It is noticed that the difference between the shNC and the USP35 one is not particularly significant in vivo, suggesting that the effect of silencing USP35 on tumor formation might not be limited to the regulation of STING-mediated interferon signaling. Together with data showing that high expression of USP35 predicted worse prognosis in ovarian cancer, our study suggests that USP35 might be a potential therapeutic target in ovarian cancer.
Activation of STING-associated type I interferons pathway is correlated with infiltration of antitumor T cells [31,32,33]. STING agonist treatment induced increased CD8+ lymphocyte intratumoral infiltration in mice model of ovarian cancer [11]. Grabosch et al. reported that cisplatin treatment stimulated cGAS/STING pathway and increased accumulation of CD8+ T cell in ovarian cancer [58]. Moreover, the higher infiltration level of intratumoral CD8+ T cell were found in the tumors of platinum-sensitive patients and predicted favorable prognosis in ovarian cancer [62, 63]. The current findings that USP35 affects IFN-β expression by deubiquitinating STING prompted us to investigate whether USP35 has any link to CD8+ T cell infiltration. Indeed, our data from immunohistochemistry and publicly available datasets showed that there is a correlation between USP35 expression and CD8+ T cell infiltration not only in ovarian cancer, but also in several other types of cancers, suggesting that USP35 may have wider functions in cancers.
In summary, our findings identify USP35 as a negative regulator of STING-associated type I interferons signaling in ovarian cancer. By investigating the effects of USP35 on its substrate STING, we have revealed that silencing USP35 elicits potent antitumor activity associated with increased expression of type I interferons and chemokines in response to cisplatin treatment. Thus, our study indicates that USP35 may be involved in the regulation of cisplatin sensitivity, suggesting it may be exploited as a promising therapeutic target to improve prognosis of the patients with ovarian cancer.
References
Lheureux S, Gourley C, Vergote I, Oza AM. Epithelial ovarian cancer. Lancet. 2019;393:1240–53.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Shang S, Yang J, Jazaeri AA, Duval AJ, Tufan T, Lopes Fischer N, et al. Chemotherapy-induced distal enhancers drive transcriptional programs to maintain the chemoresistant state in ovarian cancer. Cancer Res. 2019;79:4599–611.
Jayson GC, Kohn EC, Kitchener HC, Ledermann JA. Ovarian cancer. Lancet. 2014;384:1376–88.
Parker RJ, Eastman A, Bostick-Bruton F, Reed E. Acquired cisplatin resistance in human ovarian cancer cells is associated with enhanced repair of cisplatin-DNA lesions and reduced drug accumulation. J Clin Invest. 1991;87:772–7.
Shen J, Zhao W, Ju Z, Wang L, Peng Y, Labrie M, et al. PARPi triggers the STING-dependent immune response and enhances the therapeutic efficacy of immune checkpoint blockade independent of BRCAness. Cancer Res. 2019;79:311–19.
Kwon J, Bakhoum SF. The cytosolic DNA-sensing cGAS-STING pathway in. Cancer Cancer Disco. 2020;10:26–39.
Motwani M, Pesiridis S, Fitzgerald KA. DNA sensing by the cGAS-STING pathway in health and disease. Nat Rev Genet. 2019;20:657–74.
Kato K, Omura H, Ishitani R, Nureki O. Cyclic GMP-AMP as an endogenous second messenger in innate immune signaling by cytosolic DNA. Annu Rev Biochem. 2017;86:541–66.
Li T, Chen ZJ. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J Exp Med. 2018;215:1287–99.
Ghaffari A, Peterson N, Khalaj K, Vitkin N, Robinson A, Francis JA, et al. STING agonist therapy in combination with PD-1 immune checkpoint blockade enhances response to carboplatin chemotherapy in high-grade serous ovarian cancer. Br J Cancer. 2018;119:440–9.
Sokolowska O, Nowis D. STING signaling in cancer cells: important or not? Arch Immunol Ther Exp (Warsz). 2018;66:125–32.
Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat Med. 2014;20:1242–53.
Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422.
Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–63.
Du Y, Lin J, Zhang R, Yang W, Quan H, Zang L, et al. Ubiquitin specific peptidase 5 promotes ovarian cancer cell proliferation through deubiquitinating HDAC2. Aging (Albany NY). 2019;11:9778–93.
Han C, Yang L, Choi HH, Baddour J, Achreja A, Liu Y, et al. Amplification of USP13 drives ovarian cancer metabolism. Nat Commun. 2016;7:13525.
Padmanabhan A, Candelaria N, Wong KK, Nikolai BC, Lonard DM, O’Malley BW, et al. USP15-dependent lysosomal pathway controls p53-R175H turnover in ovarian cancer cells. Nat Commun. 2018;9:1270.
Chen Y, Wang L, Jin J, Luan Y, Chen C, Li Y, et al. p38 inhibition provides anti-DNA virus immunity by regulation of USP21 phosphorylation and STING activation. J Exp Med. 2017;214:991–1010.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Disco. 2012;2:401–4.
Nagy Á, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73.
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110.
Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17:174.
Fang L, Teng H, Wang Y, Liao G, Weng L, Li Y, et al. SET1A-mediated mono-methylation at K342 regulates YAP activation by blocking its nuclear export and promotes tumorigenesis. Cancer Cell. 2018;34:103–18.
Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–7.
Gajewski TF, Fuertes MB, Woo SR. Innate immune sensing of cancer: clues from an identified role for type I IFNs. Cancer Immunol Immunother. 2012;61:1343–7.
Brockwell NK, Parker BS. Tumor inherent interferons: Impact on immune reactivity and immunotherapy. Cytokine. 2019;118:42–7.
Corrales L, McWhirter SM, Dubensky TW Jr, Gajewski TF. The host STING pathway at the interface of cancer and immunity. J Clin Invest. 2016;126:2404–11.
Zhu Y, An X, Zhang X, Qiao Y, Zheng T, Li X. STING: a master regulator in the cancer-immunity cycle. Mol Cancer. 2019;18:152.
Khoo LT, Chen LY. Role of the cGAS-STING pathway in cancer development and oncotherapeutic approaches. EMBO Rep. 2018;19:pii: e46935.
Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, et al. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–76.
Wang Q, Liu X, Cui Y, Tang Y, Chen W, Li S, et al. The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity. 2014;41:919–33.
Zhao B, Du F, Xu P, Shu C, Sankaran B, Bell SL, et al. A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1. Nature. 2019;569:718–22.
Zhang C, Shang G, Gui X, Zhang X, Bai XC, Chen ZJ. Structural basis of STING binding with and phosphorylation by TBK1. Nature. 2019;567:394–8.
Kipps E, Tan DS, Kaye SB. Meeting the challenge of ascites in ovarian cancer: new avenues for therapy and research. Nat Rev Cancer. 2013;13:273–82.
Parkes EE, Walker SM, Taggart LE, McCabe N, Knight LA, Wilkinson R, et al. Activation of STING-dependent innate immune signaling by S-phase-specific DNA damage in breast cancer. J Natl Cancer Inst. 2016;109. https://doi.org/10.1093/jnci/djw199.
Ding L, Kim HJ, Wang Q, Kearns M, Jiang T, Ohlson CE. et al. PARP inhibition elicits STING-dependent antitumor immunity in Brca1-deficient ovarian cancer. Cell Rep. 2018;25:2972–80.
NMGP deQueiroz, Xia T, Konno H, Barber GN. Ovarian cancer cells commonly exhibit defective STING signaling which affects sensitivity to viral oncolysis. Mol Cancer Res. 2019;17:974–86.
Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–11.
Shu HB, Wang YY. Adding to the STING. Immunity. 2014;41:871–3.
Liu Q, Wu Y, Qin Y, Hu J, Xie W, Qin FX, et al. Broad and diverse mechanisms used by deubiquitinase family members in regulating the type I interferon signaling pathway during antiviral responses. Sci Adv. 2018;4:eaar2824.
Sun H, Zhang Q, Jing YY, Zhang M, Wang HY, Cai Z, et al. USP13 negatively regulates antiviral responses by deubiquitinating STING. Nat Commun. 2017;8:15534.
Ye L, Zhang Q, Liuyu T, Xu Z, Zhang MX, Luo MH, et al. USP49 negatively regulates cellular antiviral responses via deconjugating K63-linked ubiquitination of MITA. PLoS Pathog. 2019;15:e1007680.
Zhang M, Zhang MX, Zhang Q, Zhu GF, Yuan L, Zhang DE, et al. USP18 recruits USP20 to promote innate antiviral response through deubiquitinating STING/MITA. Cell Res. 2016;26:1302–19.
Zhang L, Wei N, Cui Y, Hong Z, Liu X, Wang Q, et al. The deubiquitinase CYLD is a specific checkpoint of the STING antiviral signaling pathway. PLoS Pathog. 2018;14:e1007435.
Johns TG, Mackay IR, Callister KA, Hertzog PJ, Devenish RJ, Linnane AW. Antiproliferative potencies of interferons on melanoma cell lines and xenografts: higher efficacy of interferon beta. J Natl Cancer Inst. 1992;84(15):1185–90.
Ethiraj P, Veerappan K, Samuel S, Sivapatham S. Interferon β improves the efficacy of low dose cisplatin by inhibiting NF-κB/p-Akt signaling on HeLa cells. Biomed Pharmacother. 2016;82:124–32.
Doherty MR, Cheon H, Junk DJ, Vinayak S, Varadan V, Telli ML, et al. Interferon-beta represses cancer stem cell properties in triple-negative breast cancer. Proc Natl Acad Sci USA. 2017;114:13792–7.
Dunn GP, Bruce AT, Sheehan KC, Shankaran V, Uppaluri R, Bui JD, et al. A critical function for type I interferons in cancer immunoediting. Nat Immunol. 2005;6:722–9.
Imamura Y, Tashiro H, Tsend-Ayush G, Haruta M, Dashdemberel N, Komohara Y, et al. Novel therapeutic strategies for advanced ovarian cancer by using induced pluripotent stem cell-derived myelomonocytic cells producing interferon beta. Cancer Sci. 2018;109:3403–10.
Iwamura T, Narumi H, Suzuki T, Yanai H, Mori K, Yamashita K, et al. Novel pegylated interferon-β as strong suppressor of the malignant ascites in a peritoneal metastasis model of human cancer. Cancer Sci. 2017;108:581–9.
Rambaldi A, Introna M, Colotta F, Landolfo S, Colombo N, Mangioni C, et al. Intraperitoneal administration of interferon beta in ovarian cancer patients. Cancer. 1985;56:294–301.
White MJ, McArthur K, Metcalf D, Lane RM, Cambier JC, Herold MJ, et al. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 2014;159:1549–62.
Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162:974–86.
Grabosch S, Bulatovic M, Zeng F, Ma T, Zhang L, Ross M, et al. Cisplatin-induced immune modulation in ovarian cancer mouse models with distinct inflammation profiles. Oncogene. 2019;38:2380–93.
Pavan S, Olivero M, Corà D, Di Renzo MF. IRF-1 expression is induced by cisplatin in ovarian cancer cells and limits drug effectiveness. Eur J Cancer. 2013;49:964–73.
Ethiraj P, Veerappan K, Doraisami B, Sivapatham S. Synergistic anti-carcinogenic effect of interferon-β with cisplatin on human breast adenocarcinoma MDA MB231 cells. Int Immunopharmacol. 2014;23:222–8.
Li Q, Kawamura K, Yang S, Okamoto S, Kobayashi H, Tada Y, et al. Interferon-β produces synergistic combinatory anti-tumor effects with cisplatin or pemetrexed on mesothelioma cells. PLoS ONE. 2013;8:e72709.
Wang W, Kryczek I, Dostál L, Lin H, Tan L, Zhao L, et al. Effector T cells abrogate stroma-mediated chemoresistance in ovarian. Cancer Cell. 2016;165:1092–105.
Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci USA. 2005;102:18538–43.
Acknowledgements
This study was supported by Grants from the National Natural Science Foundation of China (81625019, 31871398, 81972676, 31870900, 31701239, 31771573, 31501135, 31701214), the Shanghai International Science and Technology Cooperation Fund Project (18410722000) the Shanghai Science and Technology Commission (19ZR1439400, 19QA1406700), the Shanghai Municipal Commission of Health and Family Planning, China (2017BR014, 2017YQ068), and Young Elite Scientist Sponsorship Program by CAST (2018QNRC001).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by A. Degterev
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, J., Chen, Y., Chen, X. et al. Deubiquitinase USP35 restrains STING-mediated interferon signaling in ovarian cancer. Cell Death Differ 28, 139–155 (2021). https://doi.org/10.1038/s41418-020-0588-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-020-0588-y
This article is cited by
-
A transcriptomic analysis in mice following a single dose of ibogaine identifies new potential therapeutic targets
Translational Psychiatry (2024)
-
USP35 promotes cell proliferation and chemotherapeutic resistance through stabilizing FUCA1 in colorectal cancer
Oncogenesis (2023)
-
Targeting USP10 induces degradation of oncogenic ANLN in esophageal squamous cell carcinoma
Cell Death & Differentiation (2023)
-
USP35 promotes HCC development by stabilizing ABHD17C and activating the PI3K/AKT signaling pathway
Cell Death Discovery (2023)
-
USP35 dimer prevents its degradation by E3 ligase CHIP through auto-deubiquitinating activity
Cellular and Molecular Life Sciences (2023)