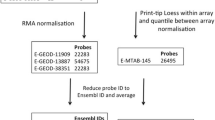
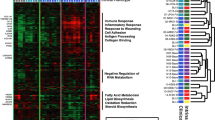
Abstract
Due to multiple hypothesis testing with often limited sample size, microarrays and other—omics technologies can sometimes produce irreproducible findings. Complementary to better experimental design, reanalysis and integration of gene expression datasets may help overcome reproducibility issues by identifying consistent differentially expressed genes from independent studies. In this work, after a systematic search, nine microarray datasets evaluating host gene expression in leprosy were reanalyzed and the information was integrated to strengthen evidence of differential expression for several genes. Our results are relevant in prioritizing genes and pathways for further investigation, whether in functional studies or in biomarker discovery. Reanalysis of individual datasets revealed several differentially expressed genes (DEGs) in accordance with original reports. Then, five integration methods (P value and effect size based) were tested. In the end, random-effects model and ratio association were selected as the main methods to pinpoint DEGs. Overall, classic pathways were found corroborating previous findings and validating this approach. Also, we identified some novel DEG involved especially with skin development processes (AQP3, AKR1C3, CYP27B1, LTB, VDR) and keratinocyte biology (CSTA, DSG1, KRT14, KRT5, PKP1, IVL), both still poorly understood in leprosy context. In addition, here we provide aggregated evidence towards some gene candidates that should be prioritized in further leprosy research, as they are likely important in immunopathogenesis. Altogether, these data are useful in better understanding host responses to the disease and, at the same time, provide a list of potential host biomarkers that could be useful in complementing leprosy diagnosis based on transcriptional levels.




Similar content being viewed by others
Data availability
All datasets analyzed during the current study are available under NCBI’s Gene Expression Omnibus repository accessions: GSE40950 (Guerreiro et al. 2013), GSE35423 (de Toledo-Pinto et al. 2016), GSE24280 [unknown], GSE443 (Bleharski et al. 2003), GSE17763 (Montoya et al. 2009), GSE16844 (Lee et al. 2010), GSE74481 (Belone et al. 2015), GSE95748 (Masaki et al. 2013), and GSE100853 (Manry et al. 2017). In addition, all R computer source code and data used in the analyzes are readily available at GitHub (https://github.com/thyagoleal/leprosy_reanalysis_paper) and Zenodo (https://doi.org/10.5281/zenodo.3840319).
Abbreviations
- sdef:
-
Ratio association method
- REM:
-
Random-effects model
- rOP :
-
r-th ordered P-value
- maxP :
-
Maximum P-value
- SR:
-
Sum of ranks
- MB:
-
Multibacillary leprosy
- PB:
-
Paucibacillary leprosy
- DEG:
-
Differentially expressed gene
- FDR:
-
False discovery rate
- BH:
-
Benjamini–Hochberg
- LL:
-
Lepromatous leprosy
- ENL:
-
Erythema nodosum leprosum
- BT:
-
Borderline-tuberculoid
- FC:
-
Fold change
- GEO:
-
Gene Expression Omnibus
- NCBI:
-
National Center for Biotechnology Information
References
Allison DB, Cui X, Page GP, Sabripour M (2006) Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet 7:55–65
Alves L, Lima LM, Silva-Maeda E, Carvalho L, Holy J, Sarno EN, Pessolani MCV, Barker LP (2004) Mycobacterium leprae infection of human Schwann cells depends on selective host kinases and pathogen-modulated endocytic pathways. FEMS Microbiol Lett 238:429–437
Alwunais KM (2015) Localized lepromatous leprosy. J Dermatol Dermatol Surg 19:133–135
Batista-Silva LR, Rodrigues LS, de Carvalho Vivarini A, Costa FD, De Mattos KA, Costa MR, Rosa PS, Toledo-Pinto TG, Dias AA, Moura DF, Sarno EN, Lopes UG, Pessolani MCV (2016) Mycobacterium leprae-induced Insulin-like Growth Factor I attenuates antimicrobial mechanisms, promoting bacterial survival in macrophages. Sci Rep 6:27632
Barbieri RR, Manta FSN, Moreira SJM, Sales AM, Nery JAC, Nascimento LPR, Hacker MA, Pacheco AG, Machado AM, Sarno EM, Moraes MO (2019) Quantitative polymerase chain reaction in paucibacillary leprosy diagnosis: a follow-up study. PLoS Negl Trop Dis 13:e0007147
Belone AD, Rosa PS, Trombone AP, Fachin LR, Guidella CC, Ura S, Barreto JA, Pinilla MG, De Carvalho AF, Carraro DM, Soares FA, Soares CT (2015) Genome-wide screening of mRNA expression in leprosy patients. Front Genet 6:1–12
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300
Blangiardo M, Richardson S (2007) Statistical tools for synthesizing lists of differentially expressed features in related experiments. Genome Biol 8:R54
Blangiardo M, Cassese A, Richardson S (2010) sdef: an R package to synthesize lists of significant features in related experiments. BMC Bioinformat 11:270
Bleharski JR, Huiying L, Meinken C, Graeber TG, Ochoa M-T, Yamamura M, Burdick A, Sarno EN, Wagner M, Rollinghoff M, Rea TH, Colonna M, Stenger S, Bloom BR, Eisenberg D, Modlin RL (2003) Use of genetic profiling in leprosy to discriminate clinical forms of the disease. Science 301(80):1527–1530
Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G (2004) GO:termFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20:3710–3715
Chang L-C, Lin H-M, Sibille E, Tseng GC (2013) Meta-analysis methods for combining multiple expression profiles: comparisons, statistical characterization and an application guideline. BMC Bioinformat 14:368
Chen JJ, Hsueh H-M, Delongchamp RR, Lin C-J, Tsai C-A (2007) Reproducibility of microarray data: a further analysis of microarray quality control (MAQC) data. BMC Bioinformat 8:412
Ching T, Huang S, Garmire LX (2014) Power analysis and sample size estimation for RNA-Seq differential expression. RNA 20:1684–1696
Choi JK, Yu U, Kim S, Yoo OJ (2003) Combining multiple microarray studies and modeling interstudy variation. Bioinformatics 19:84–90
Davis S, Meltzer PS (2007) GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 23:1846–1847
de Mattos Barbosa MG, da Silva Prata RB, Andrade PR, Ferreira H, de Andrade Silva BJ, de Oliveira JA, Assis TQ, de Toledo-Pinto TG, de Lima Bezerra OC, da Costa Nery JA, Rosa PS, Bozza MT, Lara FA, Moraes MO, Schmitz V, Sarno EN, Pinheiro RO (2017) Indoleamine 2,3-dioxygenase and iron are required for Mycobacterium leprae survival. Microbes Infect 19:505–514
de Toledo-Pinto TG, Ferreira ABR, Ribeiro-Alves M, Rodrigues LS, Batista-Silva LR, Silva BJDA, Lemes RMR, Martinez AN, Sandoval FG, Alvarado-Arnez LE, Rosa PS, Shannon EJ, Pessolani MCV, Pinheiro RO, Antunes SLG, Sarno EN, Lara FA, DiL Williams, Ozório Moraes M (2016) STING-dependent 2′-5′ oligoadenylate synthetase-like production is required for intracellular mycobacterium leprae survival. J Infect Dis 214:311–320
de Toledo-Pinto TG, Batista-Silva LR, Medeiros RCA, Lara FA, Moraes MO (2018) Type I interferons, autophagy and host metabolism in leprosy. Front Immunol 9:1–11
Desvignes LP, Ernst JD (2013) Taking sides: interferons in leprosy. Cell Host Microbe 13:377–378
Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W (2005) BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21:3439–3440
Franco-Paredes C, Rodriguez-Morales AJ (2016) Unsolved matters in leprosy: a descriptive review and call for further research. Ann Clin Microbiol Antimicrob 15:33
Gaschignard J, Grant AV, Van Thuc N, Orlova M, Cobat A, Huong NT, Ba NN, Thai VH, Abel L, Schurr E, Alcaïs A (2016) Pauci- and multibacillary leprosy: two distinct, genetically neglected diseases. PLoS Negl Trop Dis 10:1–20
Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20:307–315
Guerreiro LTA, Robottom-Ferreira AB, Ribeiro-Alves M, Toledo-Pinto TG, Rosa Brito T, Rosa PS, Sandoval FG, Jardim MR, Antunes SG, Shannon EJ, Sarno EN, Pessolani MCV, Williams DL, Moraes MO (2013) Gene expression profiling specifies chemokine, mitochondrial and lipid metabolism signatures in leprosy. PLoS ONE 8:e64748
Hedge LV, Olkin I (1985) Statistical methods for meta-analysis. Academic Press Inc, Orlando
Hernandez MDO, Fulco TDO, Pinheiro RO, Pereira RDMS, Redner P, Sarno EN, Lopes UG, Sampaio EP (2011) Thalidomide modulates Mycobacterium leprae-induced NF-κB pathway and lower cytokine response. Eur J Pharmacol 670:272–279
Hess S, Rambukkana A (2015) Bacterial-induced cell reprogramming to stem cell-like cells: new premise in host–pathogen interactions. Curr Opin Microbiol 23:179–188
Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oleś AK, Pagès H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M (2015) Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12:115–121
Ioannidis JPA, Allison DB, Ball CA, Coulibaly I, Cui X, Culhane AC, Falchi M, Furlanello C, Game L, Jurman G, Mangion J, Mehta T, Nitzberg M, Page GP, Petretto E, van Noort V (2009) Repeatability of published microarray gene expression analyses. Nat Genet 41:149–155
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264
Jacobson RR, Krahenbuhl JL (1999) Leprosy. Lancet 353:655–660
Kaur G, Kaur J (2017) Multifaceted role of lipids in Mycobacterium leprae. Future Microbiol 12:315–335
Lee DJ, Li H, Ochoa MT, Tanaka M, Carbone RJ, Damoiseaux R, Burdick A, Sarno EN, Rea TH, Modlin RL (2010) Integrated pathways for neutrophil recruitment and inflammation in leprosy. J Infect Dis 201:558–569
Lyrio ECD, Campos-Souza IC, Corrêa LCD, Lechuga GC, Verícimo M, Castro HC, Bourguignon SC, Côrte-Real S, Ratcliffe N, Declercq W, Santos DO (2015) Interaction of Mycobacterium leprae with the HaCaT human keratinocyte cell line: new frontiers in the cellular immunology of leprosy. Exp Dermatol 24:536–542
Manry J, Nédélec Y, Fava VM, Cobat A, Orlova M, Van Thuc N, Thai VH, Laval G, Barreiro LB, Schurr E (2017) Deciphering the genetic control of gene expression following Mycobacterium leprae antigen stimulation. PLoS Genet 13:e1006952
Masaki T, Qu J, Cholewa-Waclaw J, Burr K, Raaum R, Rambukkana A (2013) Reprogramming adult Schwann cells to stem cell-like cells by leprosy bacilli promotes dissemination of infection. Cell 152:51–67
Masaki T, McGlinchey A, Cholewa-Waclaw J, Qu J, Tomlinson SR, Rambukkana A (2014) Innate immune response precedes Mycobacterium leprae-induced reprogramming of adult Schwann cells. Cell Reprogram 16:9–17
Mattos KA, Lara FA, Oliveira VGC, Rodrigues LS, D’Avila H, Melo RCN, Manso PPA, Sarno EN, Bozza PT, Pessolani MCV (2011) Modulation of lipid droplets by Mycobacterium leprae in Schwann cells: a putative mechanism for host lipid acquisition and bacterial survival in phagosomes. Cell Microbiol 13:259–273
McGee M, Chen Z (2006) Parameter estimation for the exponential-normal convolution model for background correction of Affymetrix GeneChip data. Stat Appl Genet Mol Biol 5. Article 24
Montoya D, Cruz D, Teles RMB, Lee DJ, Ochoa MT, Krutzik SR, Chun R, Schenk M, Zhang X, Ferguson BG, Burdick AE, Sarno EN, Rea TH, Hewison M, Adams JS, Cheng G, Modlin RL (2009) Divergence of macrophage phagocytic and antimicrobial programs in leprosy. Cell Host Microbe 6:343–353
Moraes MO, Cardoso CC, Vanderborght PR, Pacheco AG (2006) Genetics of host response in leprosy. Lepr Rev 77:189–202
Moreau Y, Aerts S, De Moor B, De Strooper B, Dabrowski M (2003) Comparison and meta-analysis of microarray data: from the bench to the computer desk. Trends Genet 19:570–577
Phipson B, Lee S, Majewski IJ, Alexander WS, Smyth GK (2016) Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann Appl Stat 10:946–963
Ramasamy A, Mondry A, Holmes CC, Altman DG (2008) Key issues in conducting a meta-analysis of gene expression microarray datasets. PLoS Med 5:1320–1332
Rapaport F, Khanin R, Liang Y, Pirun M, Krek A, Zumbo P, Mason CE, Socci ND, Betel D (2013) Comprehensive evaluation of differential gene expression analysis methods for RNA-seq data. Genome Biol 14:R95
Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK (2007) A comparison of background correction methods for two-colour microarrays. Bioinformatics 23:2700–2707
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47
Rodrigues LC, Lockwood DNJ (2011) Leprosy now: epidemiology, progress, challenges, and research gaps. Lancet Infect Dis 11:464–470
Rung J, Brazma A (2013) Reuse of public genome-wide gene expression data. Nat Rev Genet 14:89–99
Schmitz V, Tavares IF, Pignataro P, de Machado A, dos Pacheco F, dos Santos JB, da Silva CO, Sarno EN (2019) Neutrophils in leprosy. Front Immunol 10:495
Shaw MA, Donaldson IJ, Collins A, Peacock CS, Lins-Lainson Z, Shaw JJ, Ramos F, Silveira F, Blackwell JM (2001) Association and linkage of leprosy phenotypes with HLA class II and tumour necrosis factor genes. Genes Immun 2:196–204
Shi W, Oshlack A, Smyth GK (2010) Optimizing the noise versus bias trade-off for Illumina whole genome expression BeadChips. Nucleic Acids Res 38:e204
Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:1–26
Storey JD, Tibshirani R (2003) Statistical methods for identifying differentially expressed genes in DNA microarrays. In: Brownstein MJ, Khodursky AB (eds) Functional genomics, vol 224. Methods in Molecular Biology. Humana Press
Suárez-Fariñas M, Noggle S, Heke M, Hemmati-Brivanlou A, Magnasco MO (2005) Comparing independent microarray studies: the case of human embryonic stem cells. BMC Genomics 6:99
Sweeney TE, Wong HR, Khatri P (2016) Robust classification of bacterial and viral infections via integrated host gene expression diagnostics. Sci Transl Med 8(346):346ra91. https://doi.org/10.1126/scitranslmed.aaf7165
Sweeney T, Haynes W, Vallania F, Ioannidis J, Khatri P (2017) Methods to increase reproducibility in differential gene expression via meta-analysis. Nucleic Acids Res 45(1):e1–e1
Taminau J, Lazar C, Meganck S, Nowé A (2014) Comparison of merging and meta-analysis as alternative approaches for integrative gene expression analysis. ISRN Bioinform 2014:1–7
Teles RMB, Graeber TG, Krutzik SR, Montoya D, Schenk M, Lee DJ, Komisopoulou E, Kelly-Scumpia K, Chun R, Iyer SS, Sarno EN, Rea TH, Hewison M, Adams JS, Popper SJ, Relman DA, Stenger S, Bloom BR, Cheng G, Modlin RL (2013) Type I interferon suppresses Type II interferon-triggered human anti-mycobacterial responses. Science 339(80):1448–1453
Thangaraj H, Laal S, Thangaraj I, Nath I (1988) Epidermal changes in reactional leprosy: keratinocyte Ia expression as an indicator of cell-mediated immune responses. Int J Lepr Other Mycobact Dis 56:401–407
Tseng GC, Ghosh D, Feingold E (2012) Comprehensive literature review and statistical considerations for microarray meta-analysis. Nucleic Acids Res 40:3785–3799
Wade HW (1935) Tuberculoid changes in leprosy IV. Classification of tuberculoid leprosy. Int J Lepr 3:16
Walsh CJ, Hu P, Batt J, Dos Santos CC, Ka L (2015) Microarray meta-analysis and cross-platform normalization: integrative genomics for robust biomarker discovery. Microarrays 4:389–406
Wambier C, Ramalho L, Foss N, Frade MA (2014) NF-kappa-B activation in cutaneous lesions of leprosy is associated with development of multibacillary infection. J Inflamm Res 7:133
Wan X, Pavlidis P (2007) Sharing and reusing gene expression profiling data in neuroscience. Neuroinformatics 5:161–175
Wang X, Kang DD, Shen K, Song C, Lu S, Chang LC, Liao SG, Huo Z, Tang S, Ding Y, Kaminski N, Sibille E, Lin Y, Li J, Tseng GC (2012) An r package suite for microarray meta-analysis in quality control, differentially expressed gene analysis and pathway enrichment detection. Bioinformatics 28:2534–2536
Wang Z, Arat S, Magid-Slav M, Brown JR (2018) Meta-analysis of human gene expression in response to Mycobacterium tuberculosis infection reveals potential therapeutic targets. BMC Syst Biol 12:1–18
Warsinske H, Vashisht R, Khatri P (2019) Host-response-based gene signatures for tuberculosis diagnosis: a systematic comparison of 16 signatures. PLoS Med 16:e1002786
White C, Franco-paredes C (2015) Leprosy in the 21st century. Clin Microbiol Rev 28:80–94
Yu G, Wang L-G, Han Y, He Q-Y (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. Omi A J Integr Biol 16:284–287
Acknowledgements
TLC was supported by a scholarship from the Oswaldo Cruz Institute (IOC-FIOCRUZ) from July (2016) to June (2018). We also thank the Heiser Foundation and Novartis Foundation for their financial support. The funding agencies had no involvement in the study elaboration, data analysis and interpretation or publishing process.
Author information
Authors and Affiliations
Contributions
TLC analyzed the study, interpreted data and drafted the manuscript. MOM conceptualized the study, interpreted results and reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that no competing interests exist.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors. Please refer to original reports for more information on ethical protocols adopted.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
438_2020_1705_MOESM1_ESM.zip
Supplementary Material 1—Supplementary tables S1–S4, GEO search results and detailed methods on individual study reanalysis. Full table of results for the integration/meta-analysis with FDR ≤ 0.1. This file contains four .XLS spreadsheets with results for the LL vs. Control and Stimulated vs. Control (in vitro) categories not presented within main text. One .PDF containing the 18 results from the GEOquery. One .PDF containing detailed methods used in reanalyzing each dataset individually. (ZIP 2235 kb)
438_2020_1705_MOESM2_ESM.zip
Supplementary Material 2—Supplementary Figures S1–S2, REM forest plots, multidimensional scaling (MDS) plot with top 500 most variable genes from individual datasets. Enrichment analysis for the LL vs. Control and Stimulated vs. Control (in vitro) categories and forest plots for DEGs from LL vs. BT and LL vs. ENL random effects model (REM) estimates. MDS plots from individual datasets reanalysis. (ZIP 3199 kb)
438_2020_1705_MOESM3_ESM.zip
Supplementary material 3—Supplementary Tables S5–S8 Four .XLS spreadsheets containing full enrichment results for all categories analyzed its categorical label. (ZIP 195 kb)
Rights and permissions
About this article
Cite this article
Leal-Calvo, T., Moraes, M.O. Reanalysis and integration of public microarray datasets reveals novel host genes modulated in leprosy. Mol Genet Genomics 295, 1355–1368 (2020). https://doi.org/10.1007/s00438-020-01705-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-020-01705-6