Abstract
The distinction between classification and clustering is often based on a priori knowledge of classification labels. However, in the purely theoretical situation where a data-generating model is known, the optimal solutions for clustering do not necessarily correspond to optimal solutions for classification. Exploring this divergence leads us to conclude that no standard measures of either internal or external validation can guarantee a correspondence with optimal clustering performance. We provide recommendations for the suboptimal evaluation of clustering performance. Such suboptimal approaches can provide valuable insight to researchers hoping to add a post hoc interpretation to their clusters. Indices based on pairwise linkage provide the clearest probabilistic interpretation, while a triplet-based index yields information on higher level structures in the data. Finally, a graphical examination of receiver operating characteristics generated from hierarchical clustering dendrograms can convey information that would be lost in any one number summary.






Similar content being viewed by others
References
Aidos, H., Duin, R., Fred, A. (2013). The area under the ROC curve as a criterion for clustering evaluation. In ICPRAM 2013 - proceedings of the 2nd international conference on pattern recognition applications and methods (pp. 276–280).
Albatineh, A.N., Niewiadomska-Bugaj, M., Mihalko, D. (2006). On similarity indices and correction for chance agreement. Journal of Classification, 23, 301–313.
Arbelaitz, O., Gurrutxaga, I., Muguerza, J., Pérez, J.M., Perona, I. (2013). An extensive comparative study of cluster validity indices. Pattern Recognition, 46, 243–256.
Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., Harris, M.A., Hill, D.P., Issel-Tarver, L., Kasarskis, A., Lewis, S., Matese, J.C., Richardson, J.E., Ringwald, M., Rubin, G.M., Sherlock, G. (2000). Gene ontology: tool for the unification of biology. Nature Genetics, 25(1), 25–29.
Baulieu, F. (1997). Two variant axiom systems for presence/absence based dissimilarity coefficients. Journal of Classification, 14(1), 159–170.
Baulieu, F.B. (1989). A classification of presence/absence based dissimilarity coefficients. Journal of Classification, 6(1), 233–246.
Bhattacharjee, A., Richards, W.G., Staunton, J., Li, C., Monti, S., Vasa, P., Ladd, C., Beheshti, J., Bueno, R., Gillette, M., Loda, M., Weber, G., Mark, E.J., Lander, E.S., Wong, W., Johnson, B.E., Golub, T.R., Sugarbaker, D.J., Meyerson, M. (2001). Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proceedings of the National Academy of Sciences of the United States of America, 98, 13790–13795.
Brun, M., Sima, C., Hua, J., Lowey, J., Carroll, B., Suh, E., Dougherty, E.R. (2007). Model-based evaluation of clustering validation measures. Pattern Recognition, 40(3), 807–824.
Daws, J.T. (1996). The analysis of free-sorting data: beyond pairwise cooccurrences. Journal of Classification, 13(1), 57–80.
Dougherty, E.R., & Brun, M. (2004). A probabilistic theory of clustering. Pattern Recognition, 37(5), 917–925.
Gower, J.C., & Legendre, P. (1986). Metric and Euclidean properties of dissimilarity coefficients. Journal of Classification, 3(1), 5–48.
Handl, J., Knowles, J., Kell, D.B. (2005). Computational cluster validation in post-genomic data analysis. Bioinformatics, 21(15), 3201–3212.
Hennig, C. (2015). What are the true clusters? Pattern Recognition Letters, 64, 53–62.
Hennig, C., & Liao, T.F. (2013). How to find an appropriate clustering for mixed-type variables with application to socio-economic stratification. Journal of the Royal Statistical Society: Series C (Applied Statistics), 62(3), 309–369.
Hoshida, Y., Brunet, J.P., Tamayo, P., Golub, T.R., Mesirov, J.P. (2007). Subclass mapping: identifying common subtypes in independent disease data sets. PLoS ONE, 2(11), e1195.
Hubalek, Z. (1982). Coefficients of association and similarity, based on binary (presence-absence) data: an evaluation. Biological Reviews, 57(4), 669–689.
Hubert, L., & Arabie, P. (1985). Comparing partitions. Journal of Classification, 2, 193–218.
Jain, A.K. (2010). Data clustering: 50 years beyond k-means. Pattern Recognition Letters, 31(8), 651–666.
Kaufman, L., & Rousseeuw, P.J. (Eds.). (2005). Finding groups in data: an introduction to cluster analysis. Wiley series in probability and statistics. Hoboken: Wiley.
McLachlan, G.J., & Basford, K.E. (1987). Mixture models: inference and applications to clustering. New York: Taylor & Francis.
Olsen, J.V., Vermeulen, M., Santamaria, A., Kumar, C., Miller, M.L., Jensen, L.J., Gnad, F., Cox, J., Jensen, T.S., Nigg, E.A., Brunak, S., Mann, M. (2010). Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Science Signaling, 3(104), ra3–ra3.
Qaqish, B.F., O’Brien, J.J., Hibbard, J.C., Clowers, K.J. (2017). Gene expression accelerating high-dimensional clustering with lossless data reduction. Bioinformatics, 33(18), 2867–2872.
Rand, W.M. (1971). Objective criteria for the evaluation of clustering methods. Journal of the American Statistical Association, 66, 846–850.
Rezaei, M., & Franti, P. (2016). Set matching measures for external cluster validity. IEEE Transactions on Knowledge and Data Engineering, 28(8), 2173–2186.
Seber, G.A.F. (2009). Multivariate observations. New York: Wiley.
Sing, T., Sander, O., Beerenwinkel, N., Lengauer, T. (2005). ROCR: visualizing classifier performance in R. Bioinformatics, 21(20), 3940–3941.
Thalamuthu, A., Mukhopadhyay, I., Zheng, X., Tseng, G.C. (2006). Evaluation and comparison of gene clustering methods in microarray analysis. Bioinformatics, 22 (19), 2405–2412.
Tibshirani, R., Hastie, T., Narasimhan, B., Soltys, S., Shi, G., Koong, A., Le, Q.T. (2004). Sample classification from protein mass spectrometry, by ‘Peak Probability Contrasts’. Bioinformatics, 20, 3034–3044.
Tibshirani, R., Walther, G., Hastie, T. (2001). Estimating the number of clusters in a data set via the gap statistic. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 63(2), 411–423.
Warrens, M.J. (2008a). On association coefficients for 2 × 2 tables and properties that do not depend on the marginal distributions. Psychometrika, 73(4), 777–789.
Warrens, M.J. (2008b). On the equivalence of cohen’s kappa and the Hubert-Arabie adjusted rand index. Journal of Classification, 25(2), 177–183.
Xuan Vinh, N, Julien Epps, U., Bailey, J. (2010). Information theoretic measures for clusterings comparison: variants, properties, normalization and correction for chance. Journal of Machine Learning Research, 11, 2837–2854.
Acknowledgments
The authors thank the National Cancer Institute for supporting this research through the training grant “Biostatistics for Research in Genomics and Cancer,” NCI grant 5T32CA106209-07 (T32), and the National Institute of Environmental Health Sciences for supporting it through the training grant T32ES007018.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
1.1 A.1 Proofs
Here, we present the proof from Section 2.2 that when G = 2 the clustering induced by the optimal classifier is equivalent to the optimal clustering.
We identify the two classes with the symbols 0 and 1. Any partition of the set N := {1,…,n} into two (or fewer) subsets corresponds to exactly two classifications which are mirror images of each other. The term “mirror-image” refers to flipping 0’s to 1’s and 1’s to 0’s. The optimal classifier is based on the posterior probabilities pi := P(Yi = 1|xi),i = 1,…,n. Observation i is classified as a “1” if pi > 0.5 and to “0” if pi < 0.5. The case pi = 0.5 is ignored since the xi’s are continuous (by assumption) and that event has probability 0. For a given data set X, we refer to this as the optimal classification. This optimal classification induces a partition of N. Our task is to show that the posterior probability of that partition exceeds that of any other partition (into two or fewer subsets).
Define ai = max(pi, 1 − pi) and bi = 1 − ai. The posterior probability of the optimal classification is \(A = {\prod }_{i=1}^{n} a_{i}\). The mirror-image classification has posterior probability \(B = {\prod }_{i=1}^{n} b_{i}\). Both the optimal classifier and its mirror image induce the same unique partition induced by no other classification. The posterior probability of that partition is then Q = A + B. Now we wish to prove that Q is larger than the the posterior probability of any other partition. Any alternative partition can be viewed as one induced by a modification of the optimal classification in which the classification of observation i is flipped for each i ∈ S, where S is some proper subset of N. We do not allow S = N since that leads to the same partition. We use Sc to denote the complement of S in N. Define
Note that A1 > B1 and A2 > B2 because ai > bi for each i. Now, clearly, A = A1A2,B = B1B2,Q = A1A2 + B1B2, and the posterior probability of the alternative partition is Q∗ = A1B2 + B1A2. Our aim is to prove that Q > Q∗.
Define λ = A2/(A2 + B2) and note that 1 > λ > 0.5 since A2 > B2 > 0. Since A1 > B1, we obtain the following obvious statement about convex combinations of A1 and B1,
Multiplying the leftmost and rightmost terms in the above inequality by (A2 + B2), and using the relationships λ(A2 + B2) = A2 and (1 − λ)(A2 + B2) = B2, we obtain
which is simply Q > Q∗, the desired result.
1.2 A.2 Surprising Examples
In this section, we show two examples that provide informative lessons regarding optimal criteria. Using the notation defined in Section 2, we consider a mixture of two normals; a N(0, 1) with probability π1 and a N(μ,σ2) with probability π2 = 1 − π1. The parameters are: (π1,μ,σ2). Since a linear transformation leaves the problem essentially unchanged, it suffices to consider μ > 0,σ2 ≥ 1 and π1 ∈ (0, 1).
Now we will derive optimal classifiers for two special cases, i.e., the ones that assign \(\hat {g}_{i} = \hat {g}(x_{i}) = 1\) if π1f1(xi) > π2f2(xi), and \(\hat {g}_{i} = 2\) otherwise. We will use ϕ(.,a,b) to denote the pdf of the normal distribution with mean a and variance b, and Φ() to denote the cdf of the standard normal distribution.
1.2.1 A.2.1 The Relationship Between K and G
One surprising result from our exploration is that optimal classification can occur when the number of true groups has been misspecified. Here, we show how this can occur.
Suppose σ2 > 1. The ratio
has a maximum of
In the above,
Note that M > 1 since σ2 > 1. So if M < π2/π1 then π1f1(x) < π2f2(x) and \(\hat {g}(x) = 2\) for all x.
This can arise only if π2 > 1/2. An example: π2 = 0.75,μ = 1.5,σ2 = 4,M = 2.91 < π2/π1 = 3. In this case, the optimal classifier assigns all observations to a single class, even though the model with two classes and all its parameters are completely known. It shows that accurate estimation of the number of classes is not a requirement for optimal classification.
The fact that optimal solutions can occur with the number of classes misspecified might cause some concern for researchers who design algorithms for selecting the number of classes. Popular methods for achieving this objective include the Gap Statistic (Tibshirani et al. 2001) and the Silhouette Score (Kaufman and Rousseeuw 2005).
1.2.2 A.2.2 Compactness
The above example also demonstrates that optimally assigned classes need not be compact. If M ≥ π2/π1 then \(\hat {g}(x) = 1\) if
Otherwise, we assign \(\hat {g}(x) = 2\). That is, x values within \( \sqrt { 2 \tau ^{2} \log \frac {M \pi _{1}}{\pi _{2}} } \) from 𝜃 are assigned to class 1. More extreme values, above and below 𝜃 are assigned to class 2. Hence, the region assigned to class 2 is a union of two disjoint sets. This shows that the notion that observations close together should be placed in the same class is generally false.
1.3 A.3 Theoretical Derivation of the Pairwise Indices
Now we give general expressions for the indices and the probabilities of relevant events. Let πg = P(Ai = g),g = 1,⋯ ,G, and let π denote the column vector (π1,⋯ ,πG)⊤. Of course, \({\sum }_{g=1}^{G} \pi _{g} = 1\).
For two independent observations, say observations 1 and 2, \(P(A_{1} = A_{2} ) = {\sum }_{g=1}^{G} {\pi _{g}^{2}} = \pi ^{\top } \pi = ssq(\pi )\), ssq denotes the sum of squares.
Define \(b_{gj} = P(\hat {g}_{i} = j | A_{i} = g)\) for g = 1,⋯ ,G; j = 1,⋯ ,K. The bgj’s are collected into the G × K matrix B.
The marginal distribution of \(\hat {g}_{i}\) is given by
That is, the K-vector B⊤π is the pmf of \(\hat {g}_{i}\), and
The joint probability of true linkage and its detection in a sample is
where C = diag(πg)B.
The relevant probabilities can be displayed in a 2 × 2 table as shown in Table 2. Now we easily obtain
Rand’s paremeter is γ2 + γ4. The kappa parameter is
where δ = γ1γ4 − γ2γ3.
1.4 A.4 Tables
1.4.1 A.4.1 Mixed Tumor Data
Below are the full index values for the mixed tumor example studied in Appendix. Indices are computed across clustering algorithms and the number of assumed subgroups.
CSENS | |||||||
|---|---|---|---|---|---|---|---|
K = 2 | K = 3 | K = 4 | K = 5 | K = 6 | K = 7 | k = 8 | |
K-Means | 0.98 | 0.943 | 0.802 | 0.749 | 0.724 | 0.646 | 0.625 |
Average linkage HC | 0.98 | 0.959 | 0.874 | 0.835 | 0.817 | 0.785 | 0.785 |
Single linkage HC | 0.98 | 0.959 | 0.941 | 0.884 | 0.85 | 0.85 | 0.834 |
Complete linkage HC | 0.946 | 0.843 | 0.826 | 0.756 | 0.752 | 0.653 | 0.653 |
CSPEC | |||||||
|---|---|---|---|---|---|---|---|
K = 2 | K = 3 | K = 4 | K = 5 | K = 6 | K = 7 | k = 8 | |
K-Means | 0.51 | 0.813 | 0.873 | 0.951 | 0.963 | 0.977 | 0.988 |
Average linkage HC | 0.019 | 0.038 | 0.131 | 0.166 | 0.184 | 0.715 | 0.872 |
Single linkage HC | 0.019 | 0.038 | 0.057 | 0.112 | 0.149 | 0.601 | 0.613 |
Complete linkage HC | 0.65 | 0.685 | 0.691 | 0.727 | 0.727 | 0.808 | 0.947 |
CSENS + CSPEC | |||||||
|---|---|---|---|---|---|---|---|
K = 2 | K = 3 | K = 4 | K = 5 | K = 6 | K = 7 | k = 8 | |
K-Means | 1.49 | 1.76 | 1.68 | 1.7 | 1.69 | 1.62 | 1.61 |
Average linkage HC | 1 | 0.997 | 1 | 1 | 1 | 1.5 | 1.66 |
Single linkage HC | 1 | 0.997 | 0.998 | 0.997 | 0.999 | 1.45 | 1.45 |
Complete linkage HC | 1.6 | 1.53 | 1.52 | 1.48 | 1.48 | 1.46 | 1.6 |
CPPV | |||||||
|---|---|---|---|---|---|---|---|
K = 2 | K = 3 | K = 4 | K = 5 | K = 6 | K = 7 | k = 8 | |
K-Means | 0.402 | 0.628 | 0.68 | 0.837 | 0.867 | 0.903 | 0.947 |
Average linkage HC | 0.251 | 0.251 | 0.252 | 0.252 | 0.251 | 0.48 | 0.673 |
Single linkage HC | 0.251 | 0.251 | 0.251 | 0.251 | 0.251 | 0.417 | 0.419 |
Complete linkage HC | 0.476 | 0.473 | 0.472 | 0.482 | 0.48 | 0.533 | 0.806 |
CNPV | |||||||
|---|---|---|---|---|---|---|---|
K = 2 | K = 3 | K = 4 | K = 5 | K = 6 | K = 7 | k = 8 | |
K-Means | 0.987 | 0.977 | 0.929 | 0.919 | 0.912 | 0.892 | 0.887 |
Average linkage HC | 0.748 | 0.736 | 0.756 | 0.75 | 0.75 | 0.908 | 0.923 |
Single linkage HC | 0.748 | 0.736 | 0.742 | 0.743 | 0.748 | 0.923 | 0.917 |
Complete linkage HC | 0.973 | 0.929 | 0.922 | 0.899 | 0.897 | 0.874 | 0.891 |
CPPV + CNPV | |||||||
|---|---|---|---|---|---|---|---|
K = 2 | K = 3 | K = 4 | K = 5 | K = 6 | K = 7 | k = 8 | |
K-Means | 1.39 | 1.61 | 1.61 | 1.76 | 1.78 | 1.8 | 1.83 |
Average linkage HC | 0.999 | 0.987 | 1.01 | 1 | 1 | 1.39 | 1.6 |
Single linkage HC | 0.999 | 0.987 | 0.992 | 0.994 | 0.999 | 1.34 | 1.34 |
Complete linkage HC | 1.45 | 1.4 | 1.39 | 1.38 | 1.38 | 1.41 | 1.7 |
1.4.2 A.4.2 Lung Cancer Subtype Data
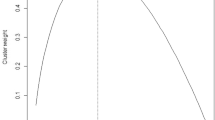
Below are the full index values for the lung cancer example studied in Appendix. Indices are computed across clustering algorithms and the number of assumed subgroups. We also show the pairwise kappa index for both datasets in Fig. 7. Notice that these plots are almost identical to the sensitivity plus specificity plots but have clear differences when compared with the triplet kappa.
Pairwise kappa for the mixed tumor dataset (Hoshida et al. 2007) (top panel) and lung cancer dataset (Bhattacharjee et al. 2001) (bottom panel). There are noticeable differences between this plot and the one presented in the paper for triplet kappa. This is highly suggestive that the triplet based index is capable of picking up information from higher order structures in the data
CSENS | |||||||
|---|---|---|---|---|---|---|---|
K = 2 | K = 3 | K = 4 | K = 5 | K = 6 | K = 7 | k = 8 | |
K-Means | 0.998 | 0.511 | 0.425 | 0.365 | 0.379 | 0.37 | 0.263 |
Average linkage HC | 1 | 0.986 | 0.984 | 0.975 | 0.962 | 0.948 | 0.947 |
Single linkage HC | 0.998 | 0.996 | 0.983 | 0.969 | 0.968 | 0.96 | 0.96 |
Complete linkage HC | 0.93 | 0.651 | 0.424 | 0.414 | 0.264 | 0.264 | 0.259 |
CSPEC | |||||||
|---|---|---|---|---|---|---|---|
K = 2 | K = 3 | K = 4 | K = 5 | K = 6 | K = 7 | k = 8 | |
K-Means | 0.351 | 0.682 | 0.81 | 0.876 | 0.94 | 0.941 | 0.957 |
Average linkage HC | 0.368 | 0.373 | 0.373 | 0.373 | 0.378 | 0.382 | 0.382 |
Single linkage HC | 0.018 | 0.035 | 0.041 | 0.047 | 0.065 | 0.205 | 0.362 |
Complete linkage HC | 0.404 | 0.688 | 0.839 | 0.845 | 0.87 | 0.878 | 0.92 |
CSENS + CSPEC | |||||||
|---|---|---|---|---|---|---|---|
K = 2 | K = 3 | K = 4 | K = 5 | K = 6 | K = 7 | k = 8 | |
K-Means | 1.35 | 1.19 | 1.23 | 1.24 | 1.32 | 1.31 | 1.22 |
Average linkage HC | 1.37 | 1.36 | 1.36 | 1.35 | 1.34 | 1.33 | 1.33 |
Single linkage HC | 1.02 | 1.03 | 1.02 | 1.02 | 1.03 | 1.16 | 1.32 |
Complete linkage HC | 1.33 | 1.34 | 1.26 | 1.26 | 1.13 | 1.14 | 1.18 |
CPPV | |||||||
|---|---|---|---|---|---|---|---|
K = 2 | K = 3 | K = 4 | K = 5 | K = 6 | K = 7 | k = 8 | |
K-Means | 0.603 | 0.614 | 0.688 | 0.744 | 0.862 | 0.862 | 0.858 |
Average linkage HC | 0.61 | 0.608 | 0.608 | 0.606 | 0.604 | 0.603 | 0.602 |
Single linkage HC | 0.501 | 0.505 | 0.503 | 0.501 | 0.506 | 0.544 | 0.598 |
Complete linkage HC | 0.607 | 0.673 | 0.722 | 0.726 | 0.667 | 0.682 | 0.761 |
CNPV | |||||||
|---|---|---|---|---|---|---|---|
K = 2 | K = 3 | K = 4 | K = 5 | K = 6 | K = 7 | k = 8 | |
K-Means | 0.993 | 0.585 | 0.587 | 0.582 | 0.605 | 0.602 | 0.568 |
Average linkage HC | 0.999 | 0.964 | 0.959 | 0.939 | 0.909 | 0.882 | 0.879 |
Single linkage HC | 0.905 | 0.907 | 0.708 | 0.609 | 0.669 | 0.84 | 0.902 |
Complete linkage HC | 0.853 | 0.666 | 0.595 | 0.593 | 0.544 | 0.547 | 0.557 |
CPPV+CNPV | |||||||
|---|---|---|---|---|---|---|---|
K = 2 | K = 3 | K = 4 | K = 5 | K = 6 | K 7 | k = 8 | |
K-Means | 1.6 | 1.2 | 1.28 | 1.33 | 1.47 | 1.46 | 1.43 |
Average linkage HC | 1.61 | 1.57 | 1.57 | 1.54 | 1.51 | 1.48 | 1.48 |
Single linkage HC | 1.41 | 1.41 | 1.21 | 1.11 | 1.17 | 1.38 | 1.5 |
Complete linkage HC | 1.46 | 1.34 | 1.32 | 1.32 | 1.21 | 1.23 | 1.32 |
1.5 6.5 Distribution of Hierarchical Clustering Assignments
The below contingency tables show distribution of subtypes in each cluster. Hierarchical clustering has a tendency to put outliers into separate clusters. Consequently, we see all of the subgroups being placed into the same cluster when K = 6. As more clusters are allowed we start to see some separation among the subtypes. This corresponds to the improved performance indices discussed in the paper.
Contingency table of cluster assignments for the mixed tumor data when K = 6
Pathology/cluster | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
Breast | 23 | 1 | 1 | 1 | 0 | 0 |
Colon | 23 | 0 | 0 | 0 | 0 | 0 |
Lung | 22 | 0 | 0 | 0 | 1 | 5 |
Prostate | 25 | 1 | 0 | 0 | 0 | 0 |
Contingency table of cluster assignments for the mixed tumor data when K = 7
Pathology/cluster | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
Breast | 21 | 1 | 1 | 2 | 1 | 0 | 0 |
Colon | 0 | 0 | 0 | 23 | 0 | 0 | 0 |
Lung | 22 | 0 | 0 | 0 | 0 | 1 | 5 |
Prostate | 0 | 1 | 0 | 25 | 0 | 0 | 0 |
Contingency table of cluster assignments for the Mixed Tumor Data when K = 8
Pathology/cluster | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
Breast | 21 | 1 | 1 | 2 | 1 | 0 | 0 | 0 |
Colon | 0 | 0 | 0 | 23 | 0 | 0 | 0 | 0 |
Lung | 22 | 0 | 0 | 0 | 0 | 0 | 1 | 5 |
Prostate | 0 | 1 | 0 | 0 | 0 | 25 | 0 | 0 |
Rights and permissions
About this article
Cite this article
O’Brien, J.J., Lawson, M.T., Schweppe, D.K. et al. Suboptimal Comparison of Partitions. J Classif 37, 435–461 (2020). https://doi.org/10.1007/s00357-019-09329-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00357-019-09329-1