Abstract
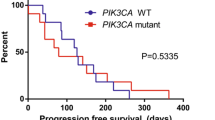
Increased expression or amplification of HER2 receptor tyrosine kinase gene ERBB2 is well-known and widely used as a prognostic biomarker of breast cancer (BC) response to the targeted treatment with trastuzumab and its analogs. Considering that part of the BC patients overexpressing HER2 does not respond to trastuzumab, clinical trial NCT03521245 was initiated to identify additional gene expression and molecular pathway activation response biomarkers to trastuzumab treatment in HER2-positive BC. Using RNA sequencing gene expression in 23 formalin-fixed, paraffin embedded HER2 positive BC tissue blocks from patients who either responded or not responded to trastuzumab treatment was profiled. Differentially regulated genes and molecular pathways were identified in the groups of trastuzumab responders and non-responders. These results were next compared with the 42 previously published BC trastuzumab responder and non-responder RNA sequencing profiles from the clinical trials NCT00513292 and NCT00353483. No correlation was observed between the response status and the expression levels of ERBB2 gene in the HER2 positive BC samples. Analysis of the differentially expressed genes and molecular pathways in the combined dataset revealed 15/27 commonly up/down regulated genes and 15/25 pathways, respectively. However, only the intersection of molecular pathways upregulated in trastuzumab responders vs non-responders was statistically significantly enriched compared to the random expectation model. A classifier built using the most significantly upregulated molecular pathway – cAMP Pathway Protein Retention – demonstrated the best performance for prediction of the HER2 positive BC response to trastuzumab for both our experimental and previously reported data. This pathway also predicted time to recurrence in the combined dataset with Log-rank p-value 0.041.






Similar content being viewed by others
Abbreviations
- AUC:
-
area under ROC-curve
- BC:
-
breast cancer
- ER:
-
estrogen receptor
- PAL:
-
pathway activation level
- PR:
-
progesterone receptor
REFERENCES
Press, M. F., Bernstein, L., Thomas, P. A., Meisner, L. F., Zhou, J. Y., Ma, Y., Hung, G., Robinson, R. A., Harris, C., El-Naggar, A., Slamon, D. J., Phillips, R. N., Ross, J. S., Wolman, S. R., and Flom, K. J. (1997) HER-2/neu gene amplification characterized by fluorescence in situ hybridization: poor prognosis in node-negative breast carcinomas, J. Clin. Oncol., 15, 2894-2904, doi: https://doi.org/10.1200/JCO.1997.15.8.2894 .
Slamon, D. J., Clark, G. M., Wong, S. G., Levin, W. J., Ullrich, A., and McGuire, W. L. (1987) Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene, Science, 235, 177-182, doi: https://doi.org/10.1126/science.3798106 .
Hammond, M. E. H., Hayes, D. F., Dowsett, M., Allred, D. C., Hagerty, K. L., et al. (2010) American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer, Arch. Pathol. Lab. Med., 134, 907-922, doi: https://doi.org/10.1043/1543-2165-134.6.907 .
Lesurf, R., Griffith, O. L., Griffith, M., Hundal, J., Trani, L., Watson, M. A., Aft, R., Ellis, M. J., Ota, D., Suman, V. J., Meric-Bernstam, F., Leitch, A. M., Boughey, J. C., Unzeitig, G., Buzdar, A. U., Hunt, K. K., and Mardis, E. R. (2017) Genomic characterization of HER2-positive breast cancer and response to neoadjuvant trastuzumab and chemotherapy-results from the ACOSOG Z1041 (Alliance) trial, Ann. Oncol., 28, 1070-1077, doi: https://doi.org/10.1093/annonc/mdx048 .
Buzdin, A., Sorokin, M., Garazha, A., Sekacheva, M., Kim, E., Zhukov, N., Wang, Y., Li, X., Kar, S., Hartmann, C., Samii, A., Giese, A., and Borisov, N. (2018) Molecular pathway activation – new type of biomarkers for tumor morphology and personalized selection of target drugs, Semin. Cancer Biol., 53, 110-124, doi: https://doi.org/10.1016/j.semcancer.2018.06.003 .
Artemov, A., Aliper, A., Korzinkin, M., Lezhnina, K., Jellen, L., Zhukov, N., Roumiantsev, S., Gaifullin, N., Zhavoronkov, A., Borisov, N., and Buzdin, A. (2015) A method for predicting target drug efficiency in cancer based on the analysis of signaling pathway activation, Oncotarget, 6, 29347-29356, doi: https://doi.org/10.18632/oncotarget.5119 .
Pagani, O., Klingbiel, D., Ruhstaller, T., Nolè, F., Eppenberger, S., et al. (2017) Do all patients with advanced HER2 positive breast cancer need upfront-chemo when receiving trastuzumab? Randomized phase III trial SAKK 22/99, Ann. Oncol., 28, 305-312, doi: https://doi.org/10.1093/annonc/mdw622 .
Schmid, S., Klingbiel, D., Aebi, S., Goldhirsch, A., Mamot, C., Munzone, E., Nolè, F., Oehlschlegel, C., Pagani, O., Pestalozzi, B., Rochlitz, C., Thürlimann, B., von Moos, R., Weder, P., Zaman, K., and Ruhstaller, T. (2019) Long-term responders to trastuzumab monotherapy in first-line HER-2+ advanced breast cancer: characteristics and survival data, BMC Cancer, 19, 902, doi: https://doi.org/10.1186/s12885-019-6105-3 .
Zhu, X., and Verma, S. (2015) Targeted therapy in HER2-positive metastatic breast cancer: a review of the literature, Curr. Oncol., 22, S19-28, doi: https://doi.org/10.3747/co.22.2363 .
Slamon, D. J., Leyland-Jones, B., Shak, S., Fuchs, H., Paton, V., Bajamonde, A., Fleming, T., Eiermann, W., Wolter, J., Pegram, M., Baselga, J., and Norton, L. (2001) Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2, N. Engl. J. Med., 344, 783-792, doi: https://doi.org/10.1056/NEJM200103153441101 .
Buzdin, A., Sorokin, M., Garazha, A., Glusker, A., Aleshin, A., Poddubskaya, E., Sekacheva, M., Kim, E., Gaifullin, N., Giese, A., Seryakov, A., Rumiantsev, P., Moshkovskii, S., and Moiseev, A. (2019) RNA sequencing for research and diagnostics in clinical oncology, Semin. Cancer Biol., 60, 311-323, doi: https://doi.org/10.1016/j.semcancer.2019.07.010 .
Artcibasova, A. V., Korzinkin, M. B., Sorokin, M. I., Shegay, P. V., Zhavoronkov, A. A., Gaifullin, N., Alekseev, B. Y., Vorobyev, N. V., Kuzmin, D. V., Kaprin, A. D., Borisov, N. M., and Buzdin, A. A. (2016) MiRImpact, a new bioinformatic method using complete microRNA expression profiles to assess their overall influence on the activity of intracellular molecular pathways, Cell Cycle, 15, 689-698, doi: https://doi.org/10.1080/15384101.2016.1147633 .
Buzdin, A. A., Prassolov, V., Zhavoronkov, A. A., and Borisov, N. M. (2017) Bioinformatics meets biomedicine: OncoFinder, a quantitative approach for interrogating molecular pathways using gene expression data, Methods Mol. Biol., 1613, 53-83, doi: https://doi.org/10.1007/978-1-4939-7027-8_4 .
Borisov, N. M., Terekhanova, N. V., Aliper, A. M., Venkova, L. S., Smirnov, P. Y., Roumiantsev, S., Korzinkin, M. B., Zhavoronkov, A. A., and Buzdin, A. A. (2014) Signaling pathways activation profiles make better markers of cancer than expression of individual genes, Oncotarget, 5, 10198-10205, doi: https://doi.org/10.18632/oncotarget.2548 .
Buzdin, A., Sorokin, M., Poddubskaya, E., and Borisov, N. (2019) High-throughput mutation data now complement transcriptomic profiling: advances in molecular pathway activation analysis approach in cancer biology, Cancer Inform., 18, 1176935119838844, doi: https://doi.org/10.1177/1176935119838844 .
Zolotovskaia, M. A., Sorokin, M. I., Roumiantsev, S. A., Borisov, N. M., and Buzdin, A. A. (2018) Pathway instability is an effective new mutation-based type of cancer biomarkers, Front. Oncol., 8, 658, doi: https://doi.org/10.3389/fonc.2018.00658 .
Lezhnina, K., Kovalchuk, O., Zhavoronkov, A. A., Korzinkin, M. B., Zabolotneva, A. A., Shegay, P. V., Sokov, D. G., Gaifullin, N. M., Rusakov, I. G., Aliper, A. M., Roumiantsev, S. A., Alekseev, B. Y., Borisov, N. M., and Buzdin, A. A. (2014) Novel robust biomarkers for human bladder cancer based on activation of intracellular signaling pathways, Oncotarget, 5, 9022-9032, doi: https://doi.org/10.18632/oncotarget.2493 .
Ozerov, I. V., Lezhnina, K. V., Izumchenko, E., Artemov, A. V., Medintsev, S., Vanhaelen, Q., Aliper, A., Vijg, J., Osipov, A. N., Labat, I., West, M. D., Buzdin, A., Cantor, C. R., Nikolsky, Y., Borisov, N., Irincheeva, I., Khokhlovich, E., Sidransky, D., Camargo, M. L., and Zhavoronkov, A. (2016) In silico Pathway Activation Network Decomposition Analysis (iPANDA) as a method for biomarker development, Nat. Commun., 7, 13427, doi: https://doi.org/10.1038/ncomms13427 .
Buzdin, A. A., Zhavoronkov, A. A., Korzinkin, M. B., Roumiantsev, S. A., Aliper, A. M., Venkova, L. S., Smirnov, P. Y., and Borisov, N. M. (2014) The OncoFinder algorithm for minimizing the errors introduced by the high-throughput methods of transcriptome analysis, Front. Mol. Biosci., 1, 8, doi: https://doi.org/10.3389/fmolb.2014.00008 .
Borisov, N., Suntsova, M., Sorokin, M., Garazha, A., Kovalchuk, O., et al. (2017) Data aggregation at the level of molecular pathways improves stability of experimental transcriptomic and proteomic data, Cell Cycle, 16, 1810-1823, doi: https://doi.org/10.1080/15384101.2017.1361068 .
Zhu, Q., Izumchenko, E., Aliper, A. M., Makarev, E., Paz, K., Buzdin, A. A., Zhavoronkov, A. A., and Sidransky, D. (2015) Pathway activation strength is a novel independent prognostic biomarker for cetuximab sensitivity in colorectal cancer patients, Hum. Genome Var., 2, 15009, doi: https://doi.org/10.1038/hgv.2015.9 .
Poddubskaya, E. V., Baranova, M. P., Allina, D. O., Sekacheva, M. I., Makovskaia, L. A., Kamashev, D. E., Suntsova, M. V., Barbara, V. S., Kochergina-Nikitskaya, I. N., and Aleshin, A. A. (2019) Personalized prescription of imatinib in recurrent granulosa cell tumor of the ovary: case report, Cold Spring Harb. Mol. Case Stud., 5, doi: https://doi.org/10.1101/mcs.a003434 .
Poddubskaya, E. V., Baranova, M. P., Allina, D. O., Smirnov, P. Y., Albert, E. A., Kirilchev, A. P., Aleshin, A. A., Sekacheva, M. I., and Suntsova, M. V. (2018) Personalized prescription of tyrosine kinase inhibitors in unresectable metastatic cholangiocarcinoma, Exp. Hematol. Oncol., 7, 21, doi: https://doi.org/10.1186/s40164-018-0113-x .
Poddubskaya, E., Bondarenko, A., Boroda, A., Zotova, E., Glusker, A., Sletina, S., Makovskaia, L., Kopylov, P., Sekacheva, M., Moisseev, A., and Baranova, M. (2019) Transcriptomics-guided personalized prescription of targeted therapeutics for metastatic ALK-positive lung cancer case following recurrence on ALK inhibitors, Front. Oncol., 9, 1026, doi: https://doi.org/10.3389/fonc.2019.01026 .
Howlader, N., Cronin, K. A., Kurian, A. W., and Andridge, R. (2018) Differences in breast cancer survival by molecular subtypes in the United States, Cancer Epidemiol. Biomarkers Prev., 27, 619-626, doi: https://doi.org/10.1158/1055-9965.EPI-17-0627 .
Prat, A., Carey, L. A., Adamo, B., Vidal, M., Tabernero, J., Cortés, J., Parker, J. S., Perou, C. M., and Baselga, J. (2014) Molecular features and survival outcomes of the intrinsic subtypes within HER2-positive breast cancer, J. Natl. Cancer Inst., 106, doi: https://doi.org/10.1093/jnci/dju152 .
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., Batut, P., Chaisson, M., and Gingeras, T. R. (2013) STAR: ultrafast universal RNA-seq aligner, Bioinformatics, 29, 15-21, doi: https://doi.org/10.1093/bioinformatics/bts635 .
Love, M. I., Huber, W., and Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol., 15, 550, doi: https://doi.org/10.1186/s13059-014-0550-8 .
Sorokin, M., Kholodenko, R., Suntsova, M., Malakhova, G., Garazha, A., Kholodenko, I., Poddubskaya, E., Lantsov, D., Stilidi, I., Arhiri, P., Osipov, A., and Buzdin, A. (2018) Oncobox bioinformatical platform for selecting potentially effective combinations of target cancer drugs using high-throughput gene expression data, Cancers, 10, doi: https://doi.org/10.3390/cancers10100365 .
Croft, D., Mundo, A. F., Haw, R., Milacic, M., Weiser, J., et al. (2014) The reactome pathway knowledgebase, Nucleic Acids Res., 42, D472-D477, doi: https://doi.org/10.1093/nar/gkt1102 .
Schaefer, C. F., Anthony, K., Krupa, S., Buchoff, J., Day, M., Hannay, T., and Buetow, K. H. (2009) PID: the Pathway Interaction Database, Nucleic Acids Res., 37, D674-D679, doi: https://doi.org/10.1093/nar/gkn653 .
Kanehisa, M. (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes, Nucleic Acids Res., 28, 27-30, doi: https://doi.org/10.1093/nar/28.1.27 .
Romero, P., Wagg, J., Green, M. L., Kaiser, D., Krummenacker, M., and Karp, P. D. (2005) Computational prediction of human metabolic pathways from the complete human genome, Genome Biol., 6, R2, doi: https://doi.org/10.1186/gb-2004-6-1-r2 .
Nishimura, D. (2001) BioCarta, Biotech Software & Internet Report, 2, 117-120, doi: https://doi.org/10.1089/152791601750294344 .
Igolkina, A. A., Zinkevich, A., Karandasheva, K. O., Popov, A. A., Selifanova, M. V., Nikolaeva, D., Tkachev, V., Penzar, D., Nikitin, D. M., and Buzdin, A. (2019) H3K4me3, H3K9ac, H3K27ac, H3K27me3 and H3K9me3 histone tags suggest distinct regulatory evolution of open and condensed chromatin landmarks, Cells, 8, doi: https://doi.org/10.3390/cells8091034 .
Marggraf, M. B., Panteleev, P. V., Emelianova, A. A., Sorokin, M. I., Bolosov, I. A., Buzdin, A. A., Kuzmin, D. V., and Ovchinnikova, T. V. (2018) Cytotoxic potential of the novel horseshoe crab peptide polyphemusin III, Mar. Drugs, 16, doi: https://doi.org/10.3390/md16120466 .
Buzdin, A., Garazha, A., Sorokin, M., Glusker, A., Aleshin, A., Allina, D., Suntsova, M., Tkachev, V., Borger, P., Borisov, N., and Gaifullin, N. (2019) RNA sequencing analysis for profiling activation of cancer-associated molecular pathways, J. Clin. Oncol., 37, e13032-e13032, doi: https://doi.org/10.1200/jco.2019.37.15_suppl.e13032 .
Green, D. M., and Swets, J. A. (1966) Signal Detection Theory and Psychophysics, Wiley, New York.
Chen, L., Zhou, Y., Tang, X., Yang, C., Tian, Y., Xie, R., Chen, T., Yang, J., Jing, M., Chen, F., Wang, C., Sun, H., and Huang, Y. (2019) EGFR mutation decreases FDG uptake in non-small cell lung cancer via the NOX4/ROS/GLUT1 axis, Int. J. Oncol., 54, 370-380, doi: https://doi.org/10.3892/ijo.2018.4626 .
Tanioka, M., Fan, C., Parker, J. S., Hoadley, K. A., Hu, Z., Li, Y., et al. (2018) Integrated analysis of RNA and DNA from the phase III trial CALGB 40601 identifies predictors of response to trastuzumab-based neoadjuvant chemotherapy in HER2-positive breast cancer, Clin. Cancer Res., 24, 5292-5304, doi: https://doi.org/10.1158/1078-0432.CCR-17-3431 .
Liu, T., Cheng, G., Kang, X., Xi, Y., Zhu, Y., Wang, K., Sun, C., Ye, J., Li, P., and Yin, H. (2018) Noninvasively evaluating the grading and IDH1 mutation status of diffuse gliomas by three-dimensional pseudo-continuous arterial spin labeling and diffusion-weighted imaging, Neuroradiology, 60, 693-702, doi: https://doi.org/10.1007/s00234-018-2021-5 .
Boyd, J. C. (1997) Mathematical tools for demonstrating the clinical usefulness of biochemical markers, Scand. J. Clin. Lab. Invest. Suppl., 227, 46-63.
Hendricks, W. P. D., Briones, N., Halperin, R. F., Facista, S., Heaton, P. R., Mahadevan, D., and Kim, S. (2019) PD-1-associated gene expression signature of neoadjuvant trastuzumab-treated tumors correlates with patient survival in HER2-positive breast cancer, Cancers, 11, doi: https://doi.org/10.3390/cancers11101566 .
Okuma, H. S., Koizumi, F., Hirakawa, A., Nakatochi, M., Komori, O., Hashimoto, J., Kodaira, M., Yunokawa, M., Yamamoto, H., Yonemori, K., Shimizu, C., Fujiwara, Y., and Tamura, K. (2016) Clinical and microarray analysis of breast cancers of all subtypes from two prospective preoperative chemotherapy studies, Br. J. Cancer, 115, 411-419, doi: https://doi.org/10.1038/bjc.2016.184 .
Popovici, V., Chen, W., Gallas, B. G., Hatzis, C., Shi, W., Samuelson, F. W., Nikolsky, Y., Tsyganova, M., Ishkin, A., Nikolskaya, T., Hess, K. R., Valero, V., Booser, D., Delorenzi, M., Hortobagyi, G. N., Shi, L., Symmans, W. F., and Pusztai, L. (2010) Effect of training-sample size and classification difficulty on the accuracy of genomic predictors, Breast Cancer Res., 12, R5, doi: https://doi.org/10.1186/bcr2468 .
Shi, L., Campbell, G., Jones, W. D., Campagne, F., Wen, Z., et al. (2010) The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models, Nat. Biotechnol., 28, 827-838, doi: https://doi.org/10.1038/nbt.1665 .
Sonnenblick, A., Brohée, S., Fumagalli, D., Rothé, F., Vincent, D., Ignatiadis, M., Desmedt, C., Salgado, R., Sirtaine, N., Loi, S., Neven, P., Loibl, S., Denkert, C., Joensuu, H., Piccart, M., and Sotiriou, C. (2015) Integrative proteomic and gene expression analysis identify potential biomarkers for adjuvant trastuzumab resistance: analysis from the Fin-her phase III randomized trial, Oncotarget, 6, 30306-30316, doi: https://doi.org/10.18632/oncotarget.5080 .
Nault, R., Fader, K. A., and Zacharewski, T. (2015) RNA-Seq versus oligonucleotide array assessment of dose-dependent TCDD-elicited hepatic gene expression in mice, BMC Genomics, 16, 373, doi: https://doi.org/10.1186/s12864-015-1527-z .
SEQC/MAQC-III Consortium (2014) A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium, Nat. Biotechnol., 32, 903-914, doi: https://doi.org/10.1038/nbt.2957 .
Suntsova, M., Gaifullin, N., Allina, D., Reshetun, A., Li, X., Mendeleeva, L., Surin, V., Sergeeva, A., Spirin, P., Prassolov, V., Morgan, A., Garazha, A., Sorokin, M., and Buzdin, A. (2019) Atlas of RNA sequencing profiles for normal human tissues, Sci. Data, 6, 36, doi: https://doi.org/10.1038/s41597-019-0043-4 .
Kumar, N., Prasad, P., Jash, E., Saini, M., Husain, A., Goldman, A., and Sehrawat, S. (2018) Insights into exchange factor directly activated by cAMP (EPAC) as potential target for cancer treatment, Mol. Cell. Biochem., 447, 77-92, doi: https://doi.org/10.1007/s11010-018-3294-z .
Capitani, M., and Sallese, M. (2009) The KDEL receptor: new functions for an old protein, FEBS Lett., 583, 3863-3871, doi: https://doi.org/10.1016/j.febslet.2009.10.053 .
Maisel, S. A., and Schroeder, J. (2019) Wrong place at the wrong time: how retrograde trafficking drives cancer metastasis through receptor mislocalization, J. Cancer Metastasis Treat., 5, 7, doi: https://doi.org/10.20517/2394-4722.2018.82 .
Valabrega, G., Montemurro, F., Sarotto, I., Petrelli, A., Rubini, P., Tacchetti, C., Aglietta, M., Comoglio, P. M., and Giordano, S. (2005) TGFα expression impairs Trastuzumab-induced HER2 downregulation, Oncogene, 24, 3002-3010, doi: https://doi.org/10.1038/sj.onc.1208478 .
Ahmad, A. (2019) Current updates on Trastuzumab resistance in HER2 overexpressing breast cancers, Adv. Exp. Med. Biol., 1152, 217-228, doi: https://doi.org/10.1007/978-3-030-20301-6_10 .
Adamczyk, A., Kruczak, A., Harazin-Lechowska, A., Ambicka, A., Grela-Wojewoda, A., Domagala-Haduch, M., Janecka-Widla, A., Majchrzyk, K., Cichocka, A., Rys, J., and Niemiec, J. (2018) Relationship between HER2 gene status and selected potential biological features related to trastuzumab resistance and its influence on survival of breast cancer patients undergoing trastuzumab adjuvant treatment, Onco Targets Ther., 11, 4525-4535, doi: https://doi.org/10.2147/OTT.S166983 .
Hanker, A. B., Garrett, J. T., Estrada, M. V., Moore, P. D., Ericsson, P. G., et al. (2017) HER2-overexpressing breast cancers amplify FGFR signaling upon acquisition of resistance to dual therapeutic blockade of HER2, Clin. Cancer Res., 23, 4323-4334, doi: https://doi.org/10.1158/1078-0432.CCR-16-2287 .
Lenz, G., Hamilton, A., Geng, S., Hong, T., Kalkum, M., Momand, J., Kane, S. E., and Huss, J. M. (2018) t-Darpp activates IGF-1R signaling to regulate glucose metabolism in Trastuzumab-resistant breast cancer cells, Clin. Cancer Res., 24, 1216-1226, doi: https://doi.org/10.1158/1078-0432.CCR-17-0824 .
Martinez, V. G., O’Neill, S., Salimu, J., Breslin, S., Clayton, A., Crown, J., and O’Driscoll, L. (2017) Resistance to HER2-targeted anti-cancer drugs is associated with immune evasion in cancer cells and their derived extracellular vesicles, Oncoimmunology, 6, e1362530, doi: https://doi.org/10.1080/2162402X.2017.1362530 .
Van Rooijen, J. M., Qiu, S. Q., Timmer-Bosscha, H., van der Vegt, B., Boers, J. E., Schroder, C. P., and de Vries, E. G. E. (2018) Androgen receptor expression inversely correlates with immune cell infiltration in human epidermal growth factor receptor 2-positive breast cancer, Eur. J. Cancer, 103, 52-60, doi: https://doi.org/10.1016/j.ejca.2018.08.001 .
Funding
This study was financially supported by the OmicsWay research program in oncology and by the Russian Science Foundation (project No. 18-15-00061, M. Su., M. So., U. V., and A. B.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
For all the biosamples, informed written consents were collected from the patients to participate in the clinical trial NCT03521245 “Molecular pathway activation markers predicting efficacy of trastuzumab therapy for HER2-positive breast cancer” and to communicate the results in this report. All procedures performed in studies involving human participants were in accordance with the ethical standards of the Karelia Republic Oncological Hospital, Petrozavodsk, and the Vitamed Oncological Clinical Center, Moscow.
The authors M. Sorokin and A. Buzdin were employed by the company OmicsWay Corp. The authors declare that this study received funding from OmicsWay Corp. The funder had the following involvement in the study: data analysis and interpretation as well as the writing of this manuscript. The funder was not involved in the study design, collection of data, decision to submit this article for publication. The remaining authors declare no conflict of interest in financial or any other sphere.
Electronic supplementary material
Fig. S1.
Expression of ERBB2 gene in the cohorts of responders and non-responders. DESeq2 normalized gene counts for ERBB2 are shown on X-axis, number of samples is shown on Y-axis.
Table S1.
Clinical and molecular annotations of HER2+ BC experimental biosamples
Table S2.
Clinical and molecular annotations of HER2+ BC literature biosamples
Table S3.
List of molecular pathways investigated in this study
Table S4.
Differential gene expression between the trastuzumab responders and non-responders in the experimental dataset (NCT03521245) and in the literature dataset dbGAP phs001291.v1.p1
Table S5.
Differentially activated molecular pathways between the trastuzumab responders and non-responders in the experimental dataset (NCT03521245) and in the literature dataset dbGAP phs001291.v1.p1
Table S6.
Number of differential genes involved in biomarker pathways
Rights and permissions
About this article
Cite this article
Sorokin, M., Ignatev, K., Barbara, V. et al. Molecular Pathway Activation Markers Are Associated with Efficacy of Trastuzumab Therapy in Metastatic HER2-Positive Breast Cancer Better than Individual Gene Expression Levels. Biochemistry Moscow 85, 758–772 (2020). https://doi.org/10.1134/S0006297920070044
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297920070044