Development and Validation of a Highly Sensitive LC-MS/MS Method for the Analysis of Bile Acids in Serum, Plasma, and Liver Tissue Samples
Abstract
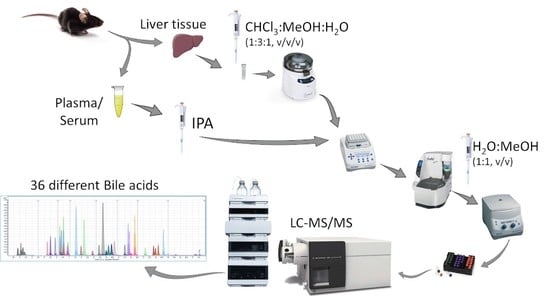
:1. Introduction
2. Results and Discussion
2.1. Establishment of the LC-MS/MS Method for Quantification of Bile Acids
2.2. Method Validation
2.3. Application of the Method
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Serum, Plasma, and Liver Tissue Samples from Mice and Plasma from Humans Used for Method Development
3.3. Mice Treated with Simvastatin
3.4. Bile Acids Extraction from Plasma and Serum
3.5. Bile Acids Extraction from Mouse Liver Tissue
3.6. Calibration and Quality Control Standard Preparation
3.7. LC-MS/MS Instrumental Conditions
3.8. Method Validation
3.9. Data Analysis and Statistics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ahmad, T.R.; Haeusler, R.A. Bile acids in glucose metabolism and insulin signalling—Mechanisms and research needs. Nat. Rev. Endocrinol. 2019, 15, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Xie, G.; Jia, W. Bile acid–microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 111–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cepa, S.; Potter, D.; Wong, L.; Schutt, L.; Tarrant, J.; Pang, J.; Zhang, X.; Andaya, R.; Salphati, L.; Ran, Y.; et al. Individual serum bile acid profiling in rats aids in human risk assessment of drug-induced liver injury due to BSEP inhibition. Toxicol. Appl. Pharm. 2018, 338, 204–213. [Google Scholar] [CrossRef]
- Slopianka, M.; Herrmann, A.; Pavkovic, M.; Ellinger-Ziegelbauer, H.; Ernst, R.; Mally, A.; Keck, M.; Riefke, B. Quantitative targeted bile acid profiling as new markers for DILI in a model of methapyrilene-induced liver injury in rats. Toxicology 2017, 386, 1–10. [Google Scholar] [CrossRef]
- Schadt, H.S.; Wolf, A.; Pognan, F.; Chibout, S.-D.D.; Merz, M.; Kullak-Ublick, G.A. Bile acids in drug induced liver injury: Key players and surrogate markers. Clin. Res. Hepatol. Gastroenterol. 2016, 40, 257–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, D.W. The Enzymes, Regulation, and Genetics of Bile Acid Synthesis. Annu. Rev. Biochem. 2003, 72, 137–174. [Google Scholar] [CrossRef] [Green Version]
- Russell, D.W.; Setchell, K.D.R. Bile Acid Biosynthesis. Biochemistry 1992, 31, 4737–4749. [Google Scholar] [CrossRef]
- Takahashi, S.; Fukami, T.; Masuo, Y.; Brocker, C.N.; Xie, C.; Krausz, K.W.; Wolf, C.R.; Henderson, C.J.; Gonzalez, F.J. Cyp2c70 is responsible for the species difference in bile acid metabolism between mice and humans. J. Lipid Res. 2016, 57, 2130–2137. [Google Scholar] [CrossRef] [Green Version]
- Tint, G.S.; Xu, G.; Batta, A.K.; Shefer, S.; Niemann, W.; Salen, G. Ursodeoxycholic acid, chenodeoxycholic acid, and 7-ketolithocholic acid are primary bile acids of the guinea pig. J. Lipid Res. 1990, 31, 1301–1306. [Google Scholar]
- Hofmann, A.F.; Hagey, L.R.; Krasowski, M.D. Bile salts of vertebrates: Structural variation and possible evolutionary significance. J. Lipid Res. 2010, 51, 226–246. [Google Scholar] [CrossRef] [Green Version]
- Monte, M.J.; Marin, J.J.; Antelo, A.; Vazquez-Tato Maria Monte, J.J.; Vazquez-Tato, J.; Chiang, J.Y.; Jjg, M. Bile acids: Chemistry, physiology, and pathophysiology. World J. Gastroenterol. 2009, 15. [Google Scholar] [CrossRef]
- Hofmann, A.F.; Mosbach, E.H. Identification of Allodeoxycholic Acid as the Major Component of Gallstones Induced in the Rabbit by 5-alpha-cholestan-3-beta-ol. J. Biol. Chem. 1964, 239, 2813–2821. [Google Scholar]
- Chiang, J.Y.L.L. Bile acids: Regulation of synthesis. J. Lipid Res. 2009, 50, 1955–1966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, J.Y.L. Bile acid metabolism and signaling. Compr. Physiol. 2013. [Google Scholar] [CrossRef] [Green Version]
- Dawson, P.A.; Karpen, S.J. Intestinal transport and metabolism of bile acids. J. Lipid Res. 2015, 56, 1085–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dawson, P.A. Role of the Intestinal Bile Acid Transporters in Bile Acid and Drug Disposition. In Handbook of Experimental Pharmacology; Springer: Berlin, Heidelberg, 2011; Volume 201, pp. 169–203. ISBN 9783642145407. [Google Scholar]
- Luo, L.; Aubrecht, J.; Li, D.; Warner, R.L.; Johnson, K.J.; Kenny, J.; Colangelo, J.L. Assessment of serum bile acid profiles as biomarkers of liver injury and liver disease in humans. PLoS ONE 2018, 13, e0193824. [Google Scholar] [CrossRef] [PubMed]
- Staley, C.; Weingarden, A.R.; Khoruts, A.; Sadowsky, M.J. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl. Microbiol. Biotechnol. 2017, 101, 47–64. [Google Scholar] [CrossRef] [Green Version]
- Urdaneta, V.; Casadesús, J. Interactions between Bacteria and Bile Salts in the Gastrointestinal and Hepatobiliary Tracts. Front. Med. 2017, 4, 1–13. [Google Scholar] [CrossRef]
- Li, T.; Apte, U. Bile Acid Metabolism and Signaling in Cholestasis, Inflammation, and Cancer. Adv. Pharmacol. 2015, 74, 263–302. [Google Scholar]
- Perez, M.J.; Britz, O. Bile-acid-induced cell injury and protection. World J. Gastroenterol. 2009, 15, 1677–1689. [Google Scholar] [CrossRef]
- Aleo, M.D.; Aubrecht, J.D.; Bonin, P.; Burt, D.A.; Colangelo, J.; Luo, L.; Schomaker, S.; Swiss, R.; Kirby, S.C.; Rigdon, G.; et al. Phase I study of PF-04895162, a Kv7 channel inhibitor, reveals unexpected hepatotoxicity in healthy subjects, but not rats or monkeys: Clinical evidence of disrupted bile acid homeostasis. Pharm. Res. Perspect. 2019, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penno, C.A.; Morgan, S.A.; Vuorinen, A.; Schuster, D.; Lavery, G.G.; Odermatt, A. Impaired oxidoreduction by 11β-hydroxysteroid dehydrogenase 1 results in the accumulation of 7-oxolithocholic acid. J. Lipid Res. 2013, 54, 2874–2883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.-M.; Liao, Y.-Y.; Lin, C.-P. Inspissated Bile Syndrome: A Rare Cause of Neonatal Cholestasis. Pediatr. Neonatol. 2020, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Clayton, P.T.; Mills, K.A.; Johnson, A.W.; Barabino, A.; Marazzi, M.G. Δ4-3-Oxosteroid 5β-reductase deficiency: Failure of ursodeoxycholic acid treatment and response to chenodeoxycholic acid plus cholic acid. Gut 1996, 38, 623–628. [Google Scholar] [CrossRef] [Green Version]
- Lemonde, H.A.; Custard, E.J.; Bouquet, J.; Duran, M.; Overmars, H.; Scambler, P.J.; Clayton, P.T. Mutations in SRD5B1 (AKR1D1), the gene encoding δ 4-3-oxosteroid 5β-reductase, in hepatitis and liver failure in infancy. Gut 2003, 52, 1494–1499. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Bathena, S.P.R.; Csanaky, I.L.; Alnouti, Y. Simultaneous characterization of bile acids and their sulfate metabolites in mouse liver, plasma, bile, and urine using LC–MS/MS. J. Pharm. Biomed. Anal. 2011, 55, 1111–1119. [Google Scholar] [CrossRef]
- Penno, C.A.; Arsenijevic, D.; Da Cunha, T.; Kullak-Ublick, G.A.; Montani, J.-P.; Odermatt, A. Quantification of multiple bile acids in uninephrectomized rats using ultra-performance liquid chromatography-tandem mass spectrometry. Anal. Methods 2013, 5, 1155. [Google Scholar] [CrossRef] [Green Version]
- Qiao, X.; Ye, M.; Liu, C.; Yang, W.; Miao, W.; Dong, J.; Guo, D. A tandem mass spectrometric study of bile acids: Interpretation of fragmentation pathways and differentiation of steroid isomers. Steroids 2012, 77, 204–211. [Google Scholar] [CrossRef]
- Lan, K.; Su, M.; Xie, G.; Ferslew, B.C.; Brouwer, K.L.R.; Rajani, C.; Liu, C.; Jia, W. Key Role for the 12-Hydroxy Group in the Negative Ion Fragmentation of Unconjugated C24 Bile Acids. Anal. Chem. 2016, 88, 7041–7048. [Google Scholar] [CrossRef] [Green Version]
- Reinicke, M.; Schröter, J.; Müller-Klieser, D.; Helmschrodt, C.; Ceglarek, U. Free oxysterols and bile acids including conjugates—Simultaneous quantification in human plasma and cerebrospinal fluid by liquid chromatography-tandem mass spectrometry. Anal. Chim. Acta 2018, 1037, 245–255. [Google Scholar] [CrossRef]
- Perwaiz, S.; Tuchweber, B.; Mignault, D.; Gilat, T.; Yousef, I.M. Determination of bile acids in biological fluids by liquid chromatography-electrospray tandem mass spectrometry. J. Lipid Res. 2001, 42, 114–119. [Google Scholar] [PubMed]
- Sarafian, M.H.; Lewis, M.R.; Pechlivanis, A.; Ralphs, S.; McPhail, M.J.W.; Patel, V.C.; Dumas, M.-E.; Holmes, E.; Nicholson, J.K. Bile Acid Profiling and Quantification in Biofluids Using Ultra-Performance Liquid Chromatography Tandem Mass Spectrometry. Anal. Chem. 2015, 87, 9662–9670. [Google Scholar] [CrossRef] [Green Version]
- García-Cañaveras, J.C.; Donato, M.T.; Castell, J.V.; Lahoz, A. Targeted profiling of circulating and hepatic bile acids in human, mouse, and rat using a UPLC-MRM-MS-validated method. J. Lipid Res. 2012, 53, 2231–2241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tagliacozzi, D.; Mozzi, A.F.; Casetta, B.; Bertucci, P.; Bernardini, S.; Di Illio, C.; Urbani, A.; Federici, G. Quantitative analysis of bile acids in human plasma by liquid chromatography-electrospray tandem mass spectrometry: A simple and rapid one-step method. Clin. Chem. Lab. Med. 2003, 41, 1633–1641. [Google Scholar] [CrossRef] [PubMed]
- Dutta, M.; Cai, J.; Gui, W.; Patterson, A.D. A review of analytical platforms for accurate bile acid measurement. Anal. Bioanal. Chem. 2019, 411, 4541–4549. [Google Scholar] [CrossRef] [PubMed]
- Ward, N.C.; Watts, G.F.; Eckel, R.H. Statin Toxicity: Mechanistic Insights and Clinical Implications. Circ. Res. 2019, 124, 328–350. [Google Scholar] [CrossRef]
- Bouitbir, J.; Sanvee, G.M.; Panajatovic, M.V.; Singh, F.; Krähenbühl, S. Mechanisms of statin-associated skeletal muscle-associated symptoms. Pharm. Res. 2020, 154, 104201. [Google Scholar] [CrossRef]
- Sanvee, G.M.; Panajatovic, M.V.; Bouitbir, J.; Krähenbühl, S. Mechanisms of insulin resistance by simvastatin in C2C12 myotubes and in mouse skeletal muscle. Biochem. Pharm. 2019, 164, 23–33. [Google Scholar] [CrossRef]
- Panajatovic, M.V.; Singh, F.; Roos, N.J.; Duthaler, U.; Handschin, C.; Krähenbühl, S.; Bouitbir, J. PGC-1α plays a pivotal role in simvastatin-induced exercise impairment in mice. Acta Physiol. 2020, 228, 1–15. [Google Scholar] [CrossRef]
- Ramesh, M.; Campos, J.C.; Lee, P.; Song, Y.; Hernandez, G.; Sin, J.; Tucker, K.C.; Saadaeijahromi, H.; Gurney, M.; Ferreira, J.C.B.; et al. Mitophagy protects against statin-mediated skeletal muscle toxicity. Faseb J. 2019, 33, 11857–11869. [Google Scholar] [CrossRef] [Green Version]
- Istvan, E.S.; Deisenhofer, J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science 2001, 292, 1160–1164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Committee for Medicinal Products for Human Use Guideline on bioanalytical method validation. Eur. Med. Agency 2011, 1–23.
- Penno, C.A.; Morgan, S.A.; Rose, A.J.; Herzig, S.; Lavery, G.G.; Odermatt, A. 11β-Hydroxysteroid dehydrogenase-1 is involved in bile acid homeostasis by modulating fatty acid transport protein-5 in the liver of mice. Mol. Metab. 2014, 3, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Taylor, F.C.; Huffman, M.; Ebrahim, S. Statin Therapy for Primary Prevention of Cardiovascular Disease. JAMA 2013, 310, 2451. [Google Scholar] [CrossRef]
- Yin, W.; Carballo-jane, E.; Mclaren, D.G.; Mendoza, V.H.; Gagen, K.; Geoghagen, N.S.; Mcnamara, L.A.; Gorski, J.N.; Eiermann, G.J.; Petrov, A.; et al. Plasma lipid profi ling across species for the identifi cation of optimal animal models of human dyslipidemia. J. Lipid Res. 2012, 53, 51–65. [Google Scholar] [CrossRef] [Green Version]
- Fu, Z.D.; Cui, J.Y.; Klaassen, C.D. Atorvastatin induces bile acid-synthetic enzyme Cyp7a1 by suppressing FXR signaling in both liver and intestine in mice. J. Lipid Res. 2014, 55, 2576–2586. [Google Scholar] [CrossRef] [Green Version]
- Fumiaki, I.; Yuri, I.; Kyoko, K.; Yutaka, K.; Yoshio, S.; Toshio, K. Effect of Simvastatin (MK-733) on Sterol and Bile Acid Excretion in Rabbits. Jpn. J. Pharm. 1990, 53, 35–45. [Google Scholar] [CrossRef] [Green Version]
- Lanzarotto, F.; Panarotto, B.; Sorbara, R.; Panteghini, M.; Pagani, F.; Sosta, S.; Lanzini, A. Effect of long term simvastatin administration as an adjunct to ursodeoxycholic acid: Evidence for a synergistic effect on biliary bile acid composition but not on serum lipids in humans. Gut 1999, 44, 552–556. [Google Scholar] [CrossRef]
- Seibert, J.; Hysek, C.M.; Penno, C.A.; Schmid, Y.; Kratschmar, D.V.; Liechti, M.E.; Odermatt, A. Acute Effects of 3,4-Methylenedioxymethamphetamine and Methylphenidate on Circulating Steroid Levels in Healthy Subjects. Neuroendocrinology 2014, 100, 17–25. [Google Scholar] [CrossRef] [Green Version]



| Compound | Abbreviation | Transition | ESI | CE (V) | RT (min) | r2 | LOD (ng/mL) | LOQ (ng/mL) | Plasma Extraction Recovery | Stability at 4 °C * | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LC (n = 6) | MC (n = 6) | HC (n = 6) | Plasma Calibration Curve (%) | Plasma Sample (%) | |||||||||
| Cholic Acid | CA | 373.3 > 355.2 | + | 8 | 9.81 | 0.999 | 0.45 | 1.36 | 111 ± 4 | 78 ± 6 | 83 ± 6 | 2.3 | 2.7 |
| Chenodeoxycholic acid | CDCA | 357.2 > 104.9 | + | 50 | 13.76 | 0.997 | 0.03 | 0.09 | 104 ± 7 | 81 ± 5 | 80 ± 6 | 0.3 | 0.5 |
| ω-muricholic acid | ωMCA | 373.3 > 159.1 | + | 20 | 6.29 | 0.999 | 0.01 | 0.02 | 113 ± 12 | 77 ± 6 | 85 ± 7 | 0.7 | 0.1 |
| α-muricholic acid | αMCA | 373.3 > 355.2 | + | 15 | 6.50 | 0.992 | 0.17 | 0.52 | 98 ± 11 | 76 ± 5 | 86 ± 10 | 1.1 | 2 |
| β-muricholic acid | βMCA | 391.3 > 355.2 | + | 16 | 6.85 | 0.997 | 0.10 | 0.30 | 94 ± 15 | 75 ± 4 | 83 ± 6 | 0.4 | 1.9 |
| γ-muricholic acid | γMCA | 373.3 > 355 | + | 10 | 8.14 | 0.991 | 0.29 | 0.86 | 87 ± 3 | 79 ± 6 | 85 ± 6 | 0.3 | ND |
| Ursodeoxycholic acid | UDCA | 357.2 > 95 | + | 35 | 9.92 | 0.997 | 0.04 | 0.12 | 94 ± 11 | 86 ± 4 | 86 ± 5 | 0.5 | ND |
| Hyodeoxycholic acid | HDCA | 357.2 > 95.1 | + | 40 | 10.34 | 0.994 | 0.41 | 1.23 | 85 ± 22 | 84 ± 4 | 88 ± 5 | 0.1 | 3.5 |
| Deoxycholic acid | DCA | 391.3 > 345 | - | 36 | 14.14 | 0.991 | 0.11 | 0.33 | 135 ± 9 | 80 ± 5 | 84 ± 5 | 1.2 | 3.9 |
| Lithocholic acid | LCA | 359.3 > 135.1 | + | 24 | 17.19 | 0.997 | 0.48 | 1.46 | <LOD | 66 ± 6 | 82 ± 5 | 1.2 | ND |
| 3-oxo-lithocholic acid | 3oxoLCA | 357.3 > 80.9 | + | 48 | 17.43 | 0.992 | 0.67 | 2.02 | <LOD | 82 ± 5 | 85 ± 7 | 3 | ND |
| 7-oxolithocholic acid | 7oxoLCA | 373.3 > 355.1 | + | 35 | 11.80 | 0.995 | 0.82 | 2.49 | 75 ± 15 | 72 ± 4 | 83 ± 6 | 1.4 | ND |
| 6,7-dioxo-lithocholic acid | 6,7dioxoLCA | 405.3 > 351.1 | + | 32 | 11.71 | 0.998 | 0.08 | 0.24 | <LOD | 1 ± 0.3 | 1 ± 0.2 | 0.7 | ND |
| 12-oxo-lithocholic acid | 12oxoLCA | 391.3 > 145.1 | + | 12 | 12.25 | 0.998 | 0.68 | 2.07 | 112 ± 13 | 68 ± 4 | 81 ± 5 | 2.5 | 0.4 |
| 7-oxo-deoxycholic acid | 7oxoDCA | 371.3 > 353.2 | + | 8 | 6.81 | 0.990 | 0.44 | 1.34 | 111 ± 15 | 83 ± 7 | 85 ± 8 | 2.3 | 2.6 |
| Allo-cholic acid | Allo-CA | 373.3 > 355.2 | + | 12 | 9.44 | 0.998 | 0.06 | 0.19 | 90 ± 9 | 77 ± 5 | 83 ± 6 | 0.3 | 1.7 |
| Allo-deoxycholic acid | Allo-DCA | 391.3 > 345 | - | 36 | 13.91 | 0.990 | 0.14 | 0.41 | 100 ± 12 | 64 ± 6 | 87 ± 6 | 0.7 | 5.9 |
| Allo-3β-deoxycholic acid | Allo-3βDCA | 391.3 > 345.1 | - | 40 | 10.10 | 0.992 | 0.51 | 1.53 | 86 ± 7 | 81 ± 5 | 83 ± 6 | 0.7 | 2.9 |
| Allo-12β-deoxycholic acid | Allo-12βDCA | 391.3 > 345.1 | - | 36 | 11.94 | 0.994 | 0.09 | 0.28 | <LOD | 80 ± 7 | 90 ± 6 | 0.2 | ND |
| Allo-3β-lithocholic acid | Allo-3βLCA | 359.3 > 135.1 | + | 25 | 15.56 | 0.987 | 0.75 | 2.26 | <LOD | 54 ± 5 | 82 ± 4 | 1.6 | ND |
| Allo-lithocholic acid | Allo-LCA | 359.3 > 135.1 | + | 25 | 17.08 | 0.995 | 0.45 | 1.37 | <LOD | 87 ± 4 | 91 ± 7 | 3.5 | ND |
| Tauro-cholic acid | TCA | 480.3 > 461.9 | + | 8 | 5.45 | 0.999 | 0.12 | 0.38 | 136 ± 15 | 82 ± 5 | 83 ± 5 | 0.3 | 2.4 |
| Tauro-chenodeoxycholic acid | TCDCA | 464.2 > 126 | + | 28 | 7.91 | 0.998 | 0.05 | 0.14 | 109 ± 8 | 80 ± 5 | 80 ± 6 | 0.1 | 2.4 |
| Tauro-ω-muricholic acid | TωMCA | 480.3 > 126 | + | 24 | 2.82 | 0.996 | 0.06 | 0.17 | 128 ± 21 | 77 ± 5 | 82 ± 5 | 0.1 | 2.7 |
| Tauro-α-muricholic acid | TαMCA | 480.3 > 126 | + | 24 | 3.07 | 0.996 | 0.06 | 0.17 | 129 ± 15 | 79 ± 6 | 81 ± 5 | 0.5 | 2.6 |
| Tauro-β-muricholic acid | TβMCA | 480.3> 126 | + | 24 | 3.30 | 0.998 | 0.04 | 0.11 | 131 ± 16 | 77 ± 4 | 78 ± 6 | 0.4 | 0.8 |
| Tauro-ursodeoxycholic acid | TUDCA | 464.2 > 126 | + | 28 | 5.20 | 0.995 | 0.03 | 0.10 | 105 ± 15 | 80 ± 5 | 83 ± 6 | 0.1 | 0.1 |
| Tauro-deoxycholic acid | TDCA | 498.2 > 124.2 | - | 45 | 8.79 | 0.995 | 0.05 | 0.14 | 117 ± 3 | 80 ± 8 | 85 ± 3 | 0.4 | 0.8 |
| Tauro-7-oxolithocholic acid | T7oxoLCA | 480.3 > 126 | + | 24 | 5.84 | 0.996 | 0.05 | 0.15 | 89 ± 5 | 78 ± 4 | 83 ± 6 | 0.5 | ND |
| Tauro-lithocholic acid | TLCA | 482.2 > 80 | - | 56 | 13.11 | 0.995 | 0.08 | 0.25 | 107 ± 5 | 70 ± 5 | 82 ± 5 | 0.9 | ND |
| Glyco-cholic acid | GCA | 464.4 >74 | - | 37 | 6.63 | 0.998 | 0.02 | 0.05 | 85 ± 5 | 81 ± 5 | 84 ± 6 | 0.4 | 2.8 |
| Glyco-chenodeoxycholic acid | GCDCA | 448.2 > 74 | - | 30 | 10.58 | 0.994 | 0.02 | 0.08 | 98 ± 5 | 82 ± 5 | 86 ± 6 | 0.8 | 0.5 |
| Glyco-ursodeoxycholic acid | GUDCA | 448.2 > 74 | - | 37 | 6.43 | 0.998 | 0.02 | 0.06 | 99 ± 12 | 80 ± 4 | 85 ± 6 | 0.3 | 3.4 |
| Glyco-deoxycholic acid | GDCA | 448.2 > 74 | - | 30 | 11.43 | 0.998 | 0.011 | 0.033 | 97 ± 9 | 81 ± 4 | 87 ± 6 | 0.4 | ND |
| Glyco-lithocholic acid | GLCA | 432.2 > 74 | - | 41 | 14.59 | 0.995 | 1.18 | 3.58 | 103 ± 13 | 70 ± 4 | 78 ± 5 | 0.1 | ND |
| Glyco-7-oxo-lithocholic acid | G7oxoLCA | 446.2 > 74 | - | 37 | 7.63 | 0.998 | 0.05 | 0.16 | 84 ± 12 | 79 ± 5 | 83 ± 6 | 1.7 | ND |
| Compound | Abbreviation | Transition | ESI | CE (V) | RT (min) | r2 | LOD (ng/mL) | LOQ (ng/mL) | Liver Extraction Recovery | Stability at 4 °C * | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LC (n = 6) | MC (n = 6) | HC (n = 6) | Liver Calibration Curve (%) | Liver Sample (%) | |||||||||
| Cholic Acid | CA | 373.3 > 355.2 | + | 8 | 9.81 | 0.999 | 0.60 | 1.81 | 103 ± 3 | 93 ± 4 | 104 ± 9 | 0.3 | 3.6 |
| Chenodeoxycholic acid | CDCA | 357.2 > 104.9 | + | 50 | 13.76 | 0.991 | 1.46 | 4.41 | <LOD | 84 ± 8 | 97 ± 5 | 0.5 | 0.4 |
| ω-muricholic acid | ωMCA | 373.3 > 159.1 | + | 20 | 6.29 | 0.998 | 0.06 | 0.18 | 105 ± 6 | 83 ± 5 | 101 ± 5 | 0.8 | 3.3 |
| α-muricholic acid | αMCA | 373.3 > 355.2 | + | 15 | 6.50 | 0.999 | 0.04 | 0.13 | 95 ± 10 | 87 ± 4 | 98 ± 3 | 0.5 | 2.3 |
| β-muricholic acid | βMCA | 391.3 > 355.2 | + | 16 | 6.85 | 0.994 | 0.32 | 0.97 | 101 ± 9 | 85 ± 5 | 103 ± 7 | 0 | 3.6 |
| γ-muricholic acid | γMCA | 373.3 > 355 | + | 10 | 8.14 | 0.997 | 0.58 | 1.76 | 87 ± 6 | 93 ± 4 | 96 ± 4 | 0.2 | ND |
| Ursodeoxycholic acid | UDCA | 357.2 > 95 | + | 35 | 9.92 | 0.995 | 0.10 | 0.30 | <LOD | 94 ± 10 | 105 ± 9 | 3.1 | 6.2 |
| Hyodeoxycholic acid | HDCA | 357.2 > 95.1 | + | 40 | 10.34 | 0.998 | 0.58 | 1.74 | <LOD | 92 ± 6 | 95 ± 6 | 0.5 | 2.6 |
| Deoxycholic acid | DCA | 391.3 > 345 | - | 36 | 14.14 | 0.996 | 0.64 | 1.94 | 88 ± 14 | 80 ± 6 | 96 ± 9 | 0.1 | 1.5 |
| Lithocholic acid | LCA | 359.3 > 135.1 | + | 24 | 17.19 | 0.979 | 1.20 | 3.64 | <LOD | 83 ± 5 | 105 ± 5 | 0.4 | ND |
| 3-oxo-lithocholic acid | 3oxoLCA | 357.3 > 80.9 | + | 48 | 17.43 | 0.994 | 0.70 | 2.13 | <LOD | 89 ± 26 | 104 ± 10 | 0.5 | ND |
| 7-oxolithocholic acid | 7oxoLCA | 373.3 > 355.1 | + | 35 | 11.80 | 0.998 | 1.07 | 3.26 | <LOD | 96 ± 4 | 106 ± 11 | 0.7 | ND |
| 12-oxo-lithocholic acid | 12oxoLCA | 391.3 > 145.1 | + | 32 | 12.25 | 0.999 | 2.55 | 7.72 | 99 ± 10 | 89 ± 4 | 99 ± 9 | 0.9 | 0.1 |
| 6,7-dioxo-lithocholic acid | 6,7dioxoLCA | 405.3 > 351.1 | + | 12 | 11.71 | 0.999 | 6.19 | 18.76 | 100 ± 10 | 86 ± 4 | 103 ± 9 | 3.1 | ND |
| 7-oxo-deoxycholic acid | 7oxoDCA | 371.3 > 353.2 | + | 8 | 6.81 | 0.995 | 0.038 | 0.115 | 92 ± 6 | 95 ± 10 | 116 ± 9 | 1 | 1.8 |
| Allo-cholic acid | Allo-CA | 373.3 > 355.2 | + | 12 | 9.44 | 0.998 | 0.14 | 0.41 | 88 ± 4 | 95 ± 4 | 105 ± 7 | 1.8 | 0.7 |
| Allo-deoxycholic acid | Allo-DCA | 391.3 > 345 | - | 36 | 13.91 | 0.995 | 0.41 | 1.23 | 93 ± 7 | 93 ± 5 | 90 ± 6 | 0.9 | ND |
| Allo-3β-deoxycholic acid | Allo-3βDCA | 391.3 > 345.1 | - | 40 | 10.10 | 0.999 | 0.36 | 1.08 | 92 ± 6 | 89 ± 4 | 97 ± 7 | 0.1 | ND |
| Allo-12β-deoxycholic acid | Allo-12βDCA | 391.3 > 345.1 | - | 36 | 11.94 | 0.991 | 0.03 | 0.10 | <LOD | 99 ± 15 | 97 ± 9 | 0.6 | ND |
| Allo-3β-lithocholic acid | Allo-3βLCA | 359.3 > 135.1 | + | 25 | 15.56 | 0.995 | 0.45 | 1.36 | <LOD | 104 ± 3 | 98 ± 4 | 1.3 | 2.4 |
| Allo-lithocholic acid | Allo-LCA | 359.3 > 135.1 | + | 25 | 17.08 | 0.997 | 0.17 | 0.52 | <LOD | 91 ± 10 | 84 ± 8 | 2.4 | ND |
| Tauro-cholic acid | TCA | 480.3 > 461.9 | + | 8 | 5.45 | 0.998 | 0.30 | 0.89 | 137 ± 10 | 88 ± 5 | 96 ± 5 | 0.5 | 3.9 |
| Tauro-chenodeoxycholic acid | TCDCA | 464.2 > 126 | + | 28 | 7.91 | 0.992 | 0.52 | 1.56 | 103 ± 15 | 84 ± 5 | 108 ± 7 | 0.6 | 0.3 |
| Tauro-ω-muricholic acid | TωMCA | 480.3 > 126 | + | 24 | 2.82 | 0.998 | 0.149 | 0.452 | 143 ± 17 | 95 ± 6 | 108 ± 5 | 0.6 | 0.9 |
| Tauro-α-muricholic acid | TαMCA | 480.3 > 126 | + | 24 | 3.07 | 0.999 | 0.10 | 0.30 | 127 ± 8 | 94 ± 6 | 106 ± 4 | 1 | 1.9 |
| Tauro-β-muricholic acid | TβMCA | 480.3> 126 | + | 24 | 3.30 | 0.999 | 0.09 | 0.27 | 189 ± 29 | 91 ± 7 | 108 ± 7 | 0 | 0.6 |
| Tauro-ursodeoxycholic acid | TUDCA | 464.2 > 126 | + | 28 | 5.20 | 0.993 | 0.45 | 1.35 | 104 ± 10 | 92 ± 6 | 101 ± 10 | 0.2 | 1.5 |
| Tauro-deoxycholic acid | TDCA | 498.2 > 124.2 | - | 45 | 8.79 | 0.991 | 0.82 | 2.48 | 78 ± 10 | 95 ± 8 | 107 ± 8 | 0.8 | 3.5 |
| Tauro-7-oxolithocholic acid | T7oxoLCA | 480.3 > 126 | + | 24 | 5.84 | 0.998 | 0.18 | 0.55 | 98 ± 6 | 87 ± 6 | 105 ± 5 | 0.2 | 4.5 |
| Tauro-lithocholic acid | TLCA | 482.2 > 80 | - | 56 | 13.11 | 0.986 | 7.21 | 21.86 | 99 ± 9 | 93 ± 6 | 91 ± 5 | 0.2 | 1.2 |
| Glyco-cholic acid | GCA | 464.4 >74 | - | 37 | 6.63 | 0.997 | 0.11 | 0.34 | 103 ± 3 | 90 ± 4 | 103 ± 3 | 0.5 | 1.4 |
| Glyco-chenodeoxycholic acid | GCDCA | 448.2 > 74 | - | 30 | 10.58 | 0.996 | 0.05 | 0.16 | 100 ± 2 | 93 ± 3 | 99 ± 6 | 2.4 | 1 |
| Glyco-ursodeoxycholic acid | GUDCA | 448.2 > 74 | - | 37 | 6.43 | 0.999 | 0.03 | 0.09 | 102 ± 6 | 91 ± 7 | 103 ± 6 | 2.7 | 1.2 |
| Glyco-deoxycholic acid | GDCA | 448.2 > 74 | - | 30 | 11.43 | 0.993 | 0.057 | 0.172 | 95 ± 3 | 89 ± 6 | 101 ± 4 | 0.5 | ND |
| Glyco-lithocholic acid | GLCA | 432.2 > 74 | - | 41 | 14.59 | 0.992 | 0.13 | 0.40 | 87 ± 5 | 94 ± 5 | 93 ± 5 | 3 | ND |
| Glyco-7-oxo-lithocholic acid | G7oxoLCA | 446.2 > 74 | - | 37 | 7.63 | 0.999 | 0.06 | 0.19 | 88 ± 4 | 91 ± 4 | 96 ± 3 | 0.4 | ND |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez, C.; Stücheli, S.; Kratschmar, D.V.; Bouitbir, J.; Odermatt, A. Development and Validation of a Highly Sensitive LC-MS/MS Method for the Analysis of Bile Acids in Serum, Plasma, and Liver Tissue Samples. Metabolites 2020, 10, 282. https://doi.org/10.3390/metabo10070282
Gómez C, Stücheli S, Kratschmar DV, Bouitbir J, Odermatt A. Development and Validation of a Highly Sensitive LC-MS/MS Method for the Analysis of Bile Acids in Serum, Plasma, and Liver Tissue Samples. Metabolites. 2020; 10(7):282. https://doi.org/10.3390/metabo10070282
Chicago/Turabian StyleGómez, Cristina, Simon Stücheli, Denise V. Kratschmar, Jamal Bouitbir, and Alex Odermatt. 2020. "Development and Validation of a Highly Sensitive LC-MS/MS Method for the Analysis of Bile Acids in Serum, Plasma, and Liver Tissue Samples" Metabolites 10, no. 7: 282. https://doi.org/10.3390/metabo10070282