Abstract
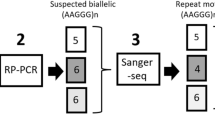
Friedreich’s ataxia (FRDA) is usually due to a homozygous GAA expansion in intron 1 of the frataxin (FXN) gene. Rarely, uncommon molecular rearrangements at the FXN locus can cause pitfalls in the molecular diagnosis of FRDA. Here we describe a family whose proband was affected by late-onset Friedreich’s ataxia (LOFA); long-range PCR (LR-PCR) documented two small expanded GAA alleles both in the proband and in her unaffected younger sister, who therefore received a diagnosis of pre-symptomatic LOFA. Later studies, however, revealed that the proband’s unaffected sister, as well as their healthy mother, were both carriers of an expanded GAA allele and an uncommon (GAAGGA)66–67 repeat mimicking a GAA expansion at the LR-PCR that was the cause of the wrong initial diagnosis of pre-symptomatic LOFA. Extensive studies in tissues from all the family members, including LR-PCR, assessment of methylation status of FXN locus, MboII restriction analysis and direct sequencing of LR-PCR products, analysis of FXN mRNA, and frataxin protein expression, support the virtual lack of pathogenicity of the rare (GAAGGA)66–67 repeat, also providing significant data about the modulation of epigenetic modifications at the FXN locus. Overall, this report highlights a rare but possible pitfall in FRDA molecular diagnosis, emphasizing the need of further analysis in case of discrepancy between clinical and molecular data.




Similar content being viewed by others
Data availability
The data that support the findings of this study are openly available in ClinVar-NCBI at the following URL: https.//www.ncbi.nlm.nih.gov/clinvar/variation/ VCV000804267.1.
References
Cossée M, Dürr A, Schmitt M et al (1999) Friedreich’s ataxia: point mutations and clinical presentation of compound heterozygotes. Ann Neurol 45:200–206
Koeppen AH, Michael SC, Knutson MD et al (2007) The dentate nucleus in Friedreich’s ataxia: the role of iron-responsive proteins. Acta Neuropathol 114(2):163–173
De Biase I, Rasmussen A, Endres D et al (2007) Progressive GAA expansions in dorsal root ganglia of Friedreich’s ataxia patients. Ann Neurol 61(1):55–60
Campuzano V, Montermini L, Lutz Y et al (1997) Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum Mol Gen 6(11):1771–1780
Filla A, De Michele G, Cavalcanti F et al (1996) The relationship between trinucleotide (GAA) repeat length and clinical features in Friedreich ataxia. Am J Hum Genet 59(3):554–560
Santoro L, De Michele G, Perretti A et al (1999) Relation between trinucleotide GAA repeat length and sensory neuropathy in Friedreich’s ataxia. J Neurol Neurosurg Psychiatry 66(1):93–96
Bit-Avragim N, Perrot A, Schöls L et al (2001) The GAA repeat expansion in intron 1 of the frataxin gene is related to the severity of cardiac manifestation in patients with Friedreich’s ataxia. J Mol Med (Berl) 78(11):626–632
Heidenfelder BL, Makhov AM, Topal MD (2003) Hairpin formation in Friedreich’s ataxia triplet repeat expansion. J Biol Chem 278(4):2425–2431
Gadgil R, Barthelemy J, Lewis T, Leffak M (2017) Replication stalling and DNA microsatellite instability. Biophys Chem 225:38–48. https://doi.org/10.1016/j.bpc.2016.11.007
Montermini L, Andermann E, Labuda M et al (1997) The Friedreich ataxia GAA triplet repeat: premutation and normal alleles. Hum Mol Genet 6(8):1261–1266
Sharma R, De Biase I, Gómez M et al (2004) Friedreich ataxia in carriers of unstable borderline GAA triplet-repeat alleles. Ann Neurol 56(6):898–901
Lazaropoulos M, Dong Y, Clark E et al (2015) Frataxin levels in peripheral tissue in Friedreich ataxia. Ann Clin Transl Neurol 2(8):831–842. https://doi.org/10.1002/acn3.225
Long A, Napierala JS, Polak U et al (2017) Somatic instability of the expanded GAA repeats in Friedreich’s ataxia. PLoS One 12(12):e0189990. https://doi.org/10.1371/journal.pone.0189990
McGinty RJ, Puleo F, Aksenova AY et al (2017) A defective mRNA cleavage and polyadenylation complex facilitates expansions of transcribed (GAA)n repeats associated with Friedreich’s ataxia. Cell Rep 20(10):2490–2500. https://doi.org/10.1016/j.celrep.2017.08.051
Castaldo I, Pinelli M, Monticelli A et al (2008) DNA methylation in intron 1 of the frataxin gene is related to GAA repeat length and age of onset in Friedreich ataxia patients. J Med Genet 45(12):808–812. https://doi.org/10.1136/jmg.2008.058594
Evans-Galea MV, Carrodus N, Rowley SM et al (2012) FXN methylation predicts expression and clinical outcome in Friedreich ataxia. Ann Neurol 71(4):487–497. https://doi.org/10.1002/ana.22671
Holloway TP, Rowley SM, Delatycki MB, Sarsero JP (2011) Detection of interruptions in the GAA trinucleotide repeat expansion in the FXN gene of Friedreich ataxia. Biotechniques 50(3):182–186. https://doi.org/10.2144/000113615
Al-Mahdawi S, Ging H, Bayot A et al (2018) Large interruptions of GAA repeat expansion mutations in Friedreich ataxia are very rare. Front Cell Neurosci 12:443. https://doi.org/10.3389/fncel.2018.00443
Cossée M, Schmitt M, Campuzano V et al (1997) Evolution of the Friedreich's ataxia trinucleotide repeat expansion: founder effect and premutations. Proc Natl Acad Sci U S A 94(14):7452–7457
Montermini L, Richter A, Morgan K et al (1997) Phenotypic variability in Friedreich ataxia: role of the associated GAA triplet repeat expansion. Ann Neurol 41(5):675–682
McDaniel DO, Keats B, Vedanarayanan VV, Subramony SH (2001) Sequence variation in GAA repeat expansions may cause differential phenotype display in Friedreich’s ataxia. Mov Disord 16(6):1153–1158
Ohshima K, Sakamoto N, Labuda M et al (1999) A nonpathogenic GAAGGA repeat in the Friedreich gene: implications for pathogenesis. Neurology 53(8):1854–1857
Stolle CA, Frackelton EC, McCallum J et al (2008) Novel, complex interruptions of the GAA repeat in small, expanded alleles of two affected siblings with late-onset Friedreich ataxia. Mov Disord 23(9):1303–1306. https://doi.org/10.1002/mds.22012
Greene E, Mahishi L, Entezam A, Kumari D, Usdin K (2007) Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res 35(10):3383–3390
Willis JH, Isaya G, Gakh O, Capaldi RA, Marusich MF (2008) Lateral-flow immunoassay for the frataxin protein in Friedreich’s ataxia patients and carriers. Mol Genet Metab 94(4):491–497. https://doi.org/10.1016/j.ymgme.2008.03.019
Barcia G, Rachid M, Magen M et al (2018) Pitfalls in molecular diagnosis of Friedreich ataxia. Eur J Med Genet 61(8):455–458. https://doi.org/10.1016/j.ejmg.2018.03.004
Sakamoto N, Ohshima K, Montermini L, Pandolfo M, Wells RD (2001) Sticky DNA, a self-associated complex formed at long GAA*TTC repeats in intron 1 of the frataxin gene, inhibits transcription. J Biol Chem 276(29):27171–27177
Polak U, Li Y, Butler JS, Napierala M (2016) Alleviating GAA repeat induced transcriptional silencing of the Friedreich’s ataxia gene during somatic cell reprogramming. Stem Cells Dev 25(23):1788–1800
Santoro M, Fontana L, Masciullo M et al (2015) Expansion size and presence of CCG/CTC/CGG sequence interruptions in the expanded CTG array are independently associated to hypermethylation at the DMPK locus in myotonic dystrophy type 1 (DM1). Biochim Biophys Acta 1852(12):2645–2652. https://doi.org/10.1016/j.bbadis.2015.09.007
Santoro M, Masciullo M, Pietrobono R et al (2013) Molecular, clinical, and muscle studies in myotonic dystrophy type 1 (DM1) associated with novel variant CCG expansions. J Neurol 260(5):1245–1257. https://doi.org/10.1007/s00415-012-6779-9
Saccà F, Puorro G, Antenora A et al (2011) A combined nucleic acid and protein analysis in Friedreich ataxia: implications for diagnosis, pathogenesis and clinical trial design. PLoS One 6(3):e17627. https://doi.org/10.1371/journal.pone.0017627
Acknowledgments
We thank Manuela Papacci and Andrea Sabino for the technical assistance and Roisin Sullivan for revising the English form of the manuscript.
Funding
This work was partially supported by grants to G.S. (Italian Ministry of University and research/M.I.U.R./Linea D1).
Author information
Authors and Affiliations
Contributions
Study conception and design, critical revision for intellectual content, manuscript draft: G. Silvestri, M. Santoro, A. Perna, S. Petrillo, F. Piemonte, and P. Chiurazzi.
Clinical data assessment and tissue collection: V. Riso, T. Nicoletti, A. Modoni.
Material preparation, experimental plan, and data analysis were performed by M. Santoro, A. Perna, S. Petrillo, La Rosa, Rossi, and Pomponi.
First draft and manuscript revised: G. Silvestri, M. Santoro, A. Perna, F. Piemonte, and P. Chiurazzi. All authors gave their contribution to critical revision and discussion of the study results, read, and approved the final form of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethics approval
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, and its later amendments or comparable ethical standards. Biological samples were obtained by all participants primarily for diagnostic purposes following appropriate written informed consent in accordance with the guidelines and with approval of the Local Ethical Committee.
Informed consent
A written informed consent was obtained from all the subjects included in this study.
Consent to participate
In the written informed consent, all subjects gave their consent to participate to this study.
Consent to publish
All the participants gave their consent to publish data regarding the results obtained in this study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Massimo Santoro and Alessia Perna wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
Rights and permissions
About this article
Cite this article
Santoro, M., Perna, A., La Rosa, P. et al. Compound heterozygosity for an expanded (GAA) and a (GAAGGA) repeat at FXN locus: from a diagnostic pitfall to potential clues to the pathogenesis of Friedreich ataxia. Neurogenetics 21, 279–287 (2020). https://doi.org/10.1007/s10048-020-00620-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-020-00620-7