ABHD11 Is Critical for Embryonic Stem Cell Expansion, Differentiation and Lipid Metabolic Homeostasis
- 1Laboratory of Stem Cell and Developmental Biology, Department of Histology and Embryology, College of Basic Medical Sciences, Third Military Medical University, Chongqing, China
- 2Department of Thoracic Surgery, Southwest Hospital, First Affiliated Hospital Third Military Medical University, Chongqing, China
- 3Cardiac Surgery Department, Southwest Hospital, First Affiliated Hospital Third Military Medical University, Chongqing, China
- 4Experimental Center of Basic Medicine, College of Basic Medical Sciences, Third Military Medical University, Chongqing, China
Growing evidence supports the notion that lipid metabolism is critical for embryonic stem cell (ESC) maintenance. Recently, α/β-hydrolase domain-containing (ABHD) proteins have emerged as novel pivotal regulators in lipid synthesis or degradation while their functions in ESCs have not been investigated. In this study, we revealed the role of ABHD11 in ESC function using classical loss and gain of function experiments. Knockout of Abhd11 hampered ESC expansion and differentiation, triggering the autophagic flux and apoptosis. In contrast, Abhd11 overexpression exerted anti-apoptotic effects in ESCs. Moreover, Abhd11 knockout disturbed GSK3β/β-Catenin and ERK signaling transduction. Finally, Abhd11 knockout led to the misexpression of key metabolic enzymes related to lipid synthesis, glycolysis, and amino acid metabolism, and ABHD11 contributed to the homeostasis of lipid metabolism. These findings provide new insights into the broad role of ABHD proteins and highlight the significance of regulators of lipid metabolism in the control of stem cell function.
Introduction
Embryonic stem cells (ESCs) are derived from the inner cell mass of early preimplantation embryo and can proliferate unlimitedly while maintaining the potential to differentiate into all somatic lineages (Nichols and Smith, 2011). Due to their self-renewal ability and pluripotency, ESCs represent a cell model for recapitulating and investigating the developmental processes, and offer unique opportunities in regenerative medicine (Wu and Belmonte, 2016). Over the past two decades, research efforts focusing on ESC biology have led to identify an orchestrated regulation involving epigenetic, transcriptional, and signaling networks, promoting pluripotency during self-renewal (Ng and Surani, 2011; Li et al., 2012; Hassani et al., 2014). Recently, growing evidences have shown that the metabolic state of ESCs is an emerging indicator of pluripotency and self-renewal (Zhang et al., 2012; Ryall et al., 2015; Sperber et al., 2015; Teslaa and Teitell, 2015). A molecular understanding of cellular metabolic homeostasis during self-renewal is essential for harnessing the full potential of ESCs.
It has been established that glucose and amino acid metabolism, along with transcriptional and epigenetic regulation, are critical for ESC pluripotency and reprogramming. For instance, glycolysis, which mediates changes in acetyl-CoA and histone acetylation, controls ESC proliferation, pluripotency, and differentiation (Kondoh et al., 2007; Moussaieff et al., 2015; Gu et al., 2016). Threonine and methionine metabolism regulates pluripotency by affecting histone methylation (Wang et al., 2009; Shyh-Chang et al., 2013; Shiraki et al., 2014). Nevertheless, the role of lipid metabolism in the regulation of self-renewal and pluripotency has been poorly investigated at the molecular level. Recent studies have revealed that the state of lipid metabolism is tightly related to the maintenance of ESC identity (Yanes et al., 2010; Zhang H. et al., 2016; Wang et al., 2017; Cornacchia et al., 2019). ESCs are characterized by the presence of abundant metabolites, including free fatty acids and secondary lipid messengers (Yanes et al., 2010). Moreover, both the lipid profile and the expression of enzymes responsible for lipid synthesis change significantly during the course of ESC differentiation, as well as during somatic cell reprogramming to pluripotency (Panopoulos et al., 2012; Wang et al., 2017). Furthermore, lipid supplements in culture medium, such as oleic acid (Wang et al., 2017) and albumin-associated lipids (Cornacchia et al., 2019) have been shown to regulate ESC pluripotency, proliferation or differentiation. However, although the lipogenic requirements for ESC pluripotency and proliferative behavior have recently been established, how the molecules implicated in lipid metabolism regulate ESC self-renewal and differentiation is still largely unclear.
Most recently, the alpha/beta hydrolase domain (ABHD) protein family members have emerged as novel pivotal regulators of lipid metabolism and signal transduction, playing roles in metabolic disease and cancers (reviewed in Lord et al., 2013). The ABHD protein family comprises more than 19 proteins, most of which have a conserved GXSXG lipase motif, predicting their possible role in lipid synthesis or degradation (reviewed in Lord et al., 2013). For instance, ABHD5 is a highly conserved regulator of lipolysis (Lass et al., 2006), while ABHD6, a monoacylglycerol hydrolase, is a critical regulator of de novo fatty acid synthesis (Thomas et al., 2013). In addition to their roles in lipid metabolism, ABHD proteins exhibit distinct functions in cell proliferation. For example, ABHD5 plays a critical role in the induction of autophagy and apoptosis (Peng et al., 2016), while ABHD2, a triacylglycerol lipase (M et al., 2016), promotes prostate cancer cell proliferation and migration (Obinata et al., 2016). However, although recent research has greatly improved our fundamental understanding of ABHD proteins in lipid metabolism and cell biology, the biochemical and physiological functions of the majority of these proteins in ESCs are still largely unknown.
In this study, we uncovered the existence of biological roles of ABHD11 in the maintenance of mouse ESCs. Our findings that ABHD11 functions as a key regulator in lipid metabolism and is also required for the expansion and differentiation of ESCs provide deeper insights into the involvement of lipid metabolism in the regulation of ESC function and differentiation.
Materials and Methods
Plasmids Construction and Transfection
CRISPR/Cas9 was applied for the knock-in of tTR-KRAB inserts into Rosa26 of R1 ESCs (Chu et al., 2016). The donor vector (pDonor-R26-tTR-KRAB-2AN) was generated by inserting a cassette of tetR-KRAB-2A-NeoR into Ai9 (Addgene, #22799) vector. The sgRNA sequence (CAGTCTTTCTAGAAGATGGG) directing a cut at 1219 bp upstream of the Rosa26 transcription start site was inserted into the CRISPR plasmid PX330 (Addgene, #42230). The tetO-CAG-Abhd11-RFP-IRES-HygroR cassette was cloned into the pPyCAGIP vector (a gift from Ian Chambers). The sgRNA sequence (TGTCTCCCAGCCAGATGTTG) targeting Abhd11 was cloned into the pLentiGuide-Puro vector (Addgene, #52963) or the pLentiCRISPR v2 vector via BsmBI restriction enzyme sites (a gift from Feng Zhang, Addgene, #52961). The pLenti-Cas9-Blast vector was obtained from Addgene (#52962). The GFP-LC3 plasmid was a gift from Dr. Hong Zheng. All plasmids and construction details are available on request. The plasmid DNA was transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Cell Culture
R1 murine ESCs were maintained as previously described (Zhang et al., 2014). Briefly, ESCs were plated on 0.2% gelatin-coated plates in ESC maintenance medium containing high glucose Dulbecco-modified Eagle medium (DMEM), 5% fetal bovine serum (FBS), 15% KnockOut Serum Replacement (KSR), 2 mM Glutamax, 1 mM sodium pyruvate, 0.1 mM NEAA, 0.1 mM β-mercaptoethanol, 100 U/ml penicillin, 100 U/ml streptomycin (all from Invitrogen) and 10 ng/ml LIF (Millipore). Cells were routinely propagated by trypsinization and replated every 2 to 3 days, with a split ratio of 1:10. For ERK inhibition, 1 μM PD0325901 was added in the maintenance medium. For 2I/LIF condition culture, ESCs were split onto a gelatin-coated dish in DMEM/F12/N2B27 medium supplemented with 1 μM PD0325901, 3 μM CHIR99021, 10 ng/ml LIF, 0.1 mM β-mercaptoethanol, and 1 mg/ml BSA. Mouse embryonic fibroblasts (MEFs) were cultured in medium containing DMEM, 10% FBS, 2 mM Glutamax, 0.1 mM NEAA. All cell cultures were maintained at 37°C under 5% CO2.
Cell Differentiation
For monolayer differentiation, ESCs were seeded at a density of 2 × 104 cells/cm2 in gelatin-coated plates and cultured for 3 days in the maintenance medium without LIF. For EB formation, single-cell suspensions were plated at a density of 1 × 104 cells/cm2 in ultra-low attachment dishes and cultured in DMEM containing 15% FBS, 2 mM glutamax, 1 mM sodium pyruvate, 0.1 mM NEAA, 0.1 mM β-mercaptoethanol. The medium was changed every 2 days. EBs were harvested at Day 0, Day 3, and Day 5.
Cell Proliferation and Colony Formation Assay
For proliferation assay, cells were seeded at a density of 1 × 105 cells/well in 12-well plates and passaged every 2 days. Viable cells were counted using Cell Counter (Countstar) and trypan blue. For colony formation assay, cells were seeded a density of 200 cells/well in 12-well plates and cultured for 5 days. Alkaline phosphatase activity was detected with a BCIP/NBT alkaline phosphatase detection kit as previously described (Zhang et al., 2014).
Lentiviral Production and Infection
Briefly, 293FT cells were grown to 80% confluence in DMEM/10% FBS. Then, 5.56 μg Lentiviral vector, 4.17 μg pSPAX2 and 2.78 μg pMD2G were co-transfected into 293FT cells by calcium phosphate transfection. Twelve hours after transfection, the medium was changed with DMEM/5% FBS. Two day later, viral supernatant was collected and concentrated by ultracentrifugation at 70,000 g for 2 h. For lentivirus infection, cells were then plated at a density of 1 × 104 cells in 24-well plates and viral along with polybrene (4 μg/ml; Sigma) were added. After 36 h, cells were trypsinized and replated at 1 × 104 cells per gelatin-coated 60-mm dish, and cultured in ESC medium supplemented with 1 μg/ml puromycin (Invitrogen) for 3 days.
Flow Cytometric Analysis
For cell cycle analysis, cells were washed twice with phosphate-buffered saline (PBS) and fixed in 70% ethanol at –20°C overnight. Then, the fixed cells were washed and incubated in PBS containing 50 μg/ml propidium iodide, 50 μg/ml RNase A, 0.2% Triton X-100, and 0.1 mM EDTA for 30 min on ice. For apoptosis analysis, cells were harvested and stained with Annexin V-APC and propidium iodide. Following staining, samples were analyzed using a flow cytometer (ACEA Novocyte).
Teratoma Formation and Histological Analysis
All of the animal experiments were approved by the Animal Ethical and Experimental Committee of Third Military Medical University. Teratoma formation and histological analysis was performed as described previously (Zhang et al., 2014). Briefly, 8 × 105 ESCs were injected into the posterior flanks of nude mice. The Abhd11OE ESCs and RFPOE ESCs injected mice were treated with 1 mg/ml doxycycline in their drinking water. After 4 weeks, tumors were collected and analyzed by hematoxylin-eosin staining.
RNA Isolation, Reverse Transcription, and PCR
RNA Isolation, reverse transcription, PCR and real-time PCR were performed as previously described (Ruan et al., 2017). All primers used in this study are listed in Supplementary Table S4.
Immunofluorescence and Western Blotting
Cells were fixed in 4% paraformaldehyde for 20 min at 4°C, and permeabilized with 0.1% Triton X-100 for 15 min, followed by blocking with 10% FBS/PBS for 30 min. Cells were stained with anti-β-Catenin (CST,1:200) for overnight at 4°C, followed by the rabbit IgG secondary antibodies (1:1000) and counterstained with Hoechst. Images were captured with a ZEISS 780 inverted confocal microscope. Western blotting was performed as previously described (Zhang et al., 2014). The antibodies used in this study are given in Supplementary Table S5.
Transcriptome Analysis
RNA-Seq transcriptome analysis was performed at Genminix Informatics Ltd., Co. (Shanghai, China) using Illumina HiSeq × 10 sequencing platform. The read counts for each gene were calculated, and the expression values of each gene were normalized by R package “DESeq2”. The differentially expressed genes (DEGs) were selected by the DESeq2. A p-values < 0.05 and an absolute value of the log2FC ≥ 0.5 were used as a threshold for significance. Among the DEGs, the up-regulated genes with counts less than 10 in the experimental group were removed, and the down-regulated genes with counts less than 10 in the control group were removed. The DEGs are listed in Supplementary Table S2. Heatmaps were generated by Hierarchical Clustering in Cluster 3.0 and visualised using Java Treeview. The reactome and GO enrichment analysis on DEGs were performed using g:Profiler1 and visualized using Cytoscape and Enrichment Map as previously reported (Reimand et al., 2019). The raw data are in GEO accession number GSE142067.
GFP-LC3 Detection
Cells were transfected with plasmid GFP-LC3 (a gift from Hong Zheng) and cultured on gelatin-coated glass slides. 36 h after transfection, cells were fixed with 4% paraformaldehyde for 20 min and counterstained with Hoechst. Images were captured with a ZEISS 780 inverted confocal microscope and analyzed using CellProfiler software (Klionsky et al., 2016).
Untargeted Relative Quantitative Lipidomics Assay
For sample preparation and lipid extraction, 6 × 105 cells were homogenized with 200 μL water and 240 μL methanol. Then 800 μL of methyl tert-butyl ether was added and the mixture was ultrasound 20 min at 4°C followed by sitting still for 30 min at room temperature. The solution was centrifuged at 14000 g for 15 min at 10°C and the upper organic solvent layer was obtained and dried under nitrogen. Lipid analysis by liquid chromatography-tandem mass spectrometry (LC-MS/MS) and data analyses were performed as instructions by Shanghai Applied Protein Technology. Briefly, reverse phase chromatography was selected for LC separation using CSH C18 column (1.7 μm, 2.1 × 100 mm, Waters). Mass spectra were acquired by Q-Exactive Plus in positive and negative mode, respectively. ESI parameters were optimized and preset for all measurements as follows: Source temperature, 300°C; Capillary Temp, 350°C, the ion spray voltage was set at 3000 V, S-Lens RF Level was set at 50% and the scan range of the instruments was set at m/z 200–1800. Lipid species were identified with LipidSearch software version 4.1 (Thermo ScientificTM). For data analysis, principal component analysis (PCA) and partial least-squares-discriminant analysis (PLS-DA) were performed. Fold change of the lipid content between Abhd11 KO or OE and control cells was calculated. The significant different lipid species, which were showed by volcano plots, were determined based on the combination of fold change, variable influence on projection (VIP) values (obtained from PLS-DA) and P-values (Student’s t-test) on the raw data.
Transmission Electron Microscopy
Cell samples were fixed using 2.5% glutaraldehyde in PBS at 4°C overnight, then washed twice with 0.1 M PBS and postfixed with 1% osmium tetroxide for 2 h at 4°C. Samples were dehydrated in increasing concentrations of acetone and double stained with uranyl acetate in 70% acetone at 4°C overnight. Samples were embedded, sectioned and examined using JEM-1400plus transmission electron microscopy.
Statistics
Statistical analysis was performed using the Statistical Package for Social Science. The Student’s t-test was used to analyze the statistical differences. Data were presented as mean value ± SD, and P < 0.05 was considered to be statistically significant. Each experiment was performed at least three times.
Results
ABHD11 Association With ESCs Is Revealed by Bioinformatic Analysis
To investigate whether ABHD proteins were implicated in mouse ESC maintenance, we first analyzed ESC transcriptome data (Supplementary Table S1) and a published genome-scale CRISPR-Cas9 knockout ESC dataset, in which genes essential for ESC self-renewal are ranked (Tzelepis et al., 2016). Notably, we found that Oct4 and Nanog, two core pluripotency genes (Loh et al., 2006), were expressed at high levels, and included in the top-1000 ranked genes. Among ABHD family genes, only Abhd11 was in the top-2500 ranked genes and expressed at a relatively high level, while the other members of the family were out of the top-5000 ranked genes (Supplementary Figure S1A). The promoters of genes that are required to maintain ESCs in a non-differentiated state are usually co-occupied by a couple of core pluripotency transcription factors (PTFs) (Boyer et al., 2005; Loh et al., 2006; Kim, 2008). We next analyzed the target promoters of nine PTFs in ESCs (Kim, 2008). Among the involved 797 genes, Abhd11 was the only ABHD family genes whose promoter was occupied by more than four factors (Supplementary Figure S1B). Furthermore, we analyzed the mRNA expression data of ABHD family genes in OCT4-positive (undifferentiated) and OCT4-negative (differentiated) cells derived from ESCs (Zhou et al., 2007). We found that Abhd11 and Abhd10 were downregulated, while most other ABHD family genes, such as Abhd 1, 3, 5, 6, and 7 were upregulated in OCT4-negative cells (Supplementary Figure S1C). Collectively, these data suggested that ABHD11 was involved in the maintenance of ESC identity.
Construction of ESC Lines With Conditional Abhd11 Expression
In order to assess the function of ABHD11 in ESCs, we attempted to generate Abhd11 knockout (KO) and overexpression (OE) ESC lines. To circumvent the difficulty of obtaining Abhd11-null ESCs, which may have severe growth disadvantages, we attempted to firstly generate cells with inducible overexpression (iOE) of exogenous Abhd11 using “tetR-KRAB-tetO” conditional gene expression system (Szulc et al., 2006) and then knock out the endogenous Abhd11 using CRISPR/Cas9 technology (Cong et al., 2013; Mali et al., 2013). To this end, we generated a stable tetR-KRAB-Cas9-expressing cell line (Figure 1A), which was hereafter used as a control in all experiments. Next, we obtained Abhd11iOE ESCs by transducing an inducible expression cassette of Abhd11 targeted with Red fluorescent protein coding sequence (Abhd11-RFP), into the tetR-KRAB-Cas9-expressing cells (Supplementary Figure S2A and Figure 1A). The expression of Abhd11-RFP in subcloned Abhd11iOE ESCs was induced in the presence of doxycycline (Dox) while silenced in the absence of Dox (Supplementary Figure S2A and Figures 1B,C). Finally, we generated Abhd11iOE/KO ESCs by transducing a lentivirus carrying sgRNAs mapping to the endogenous Abhd11 genomic sequence and spanning an intron-exon junction into Abhd11iOE ESCs (Supplementary Figure S2B and Figure 1A). Expression of endogenous Abhd11 in Abhd11iOE/KO ESCs was detected by reverse transcriptase-PCR (RT-PCR), using specific primers targeting the untranslated region (UTR). One of the subclones failed to express normal levels of the endogenous Abhd11 mRNA (Figure 1D). Western bot confirmed that, in the subclone, the endogenous ABHD11 protein was depleted, while exogenous ABHD11 could be efficiently switched on or off in 2 days (Figure 1E). Hence, endogenous Abhd11-depleted Abhd11iOE/KO ESCs were used as Abhd11-KO (Abhd11KO) ESCs in the absence of Dox, and as Abhd11-rescued (Abhd11RES) ESCs in the presence of 1 ng/ml Dox; Abhd11iOE ESCs were used as Abhd11-OE (Abhd11OE) ESCs in the presence of 1 μg/ml Dox.
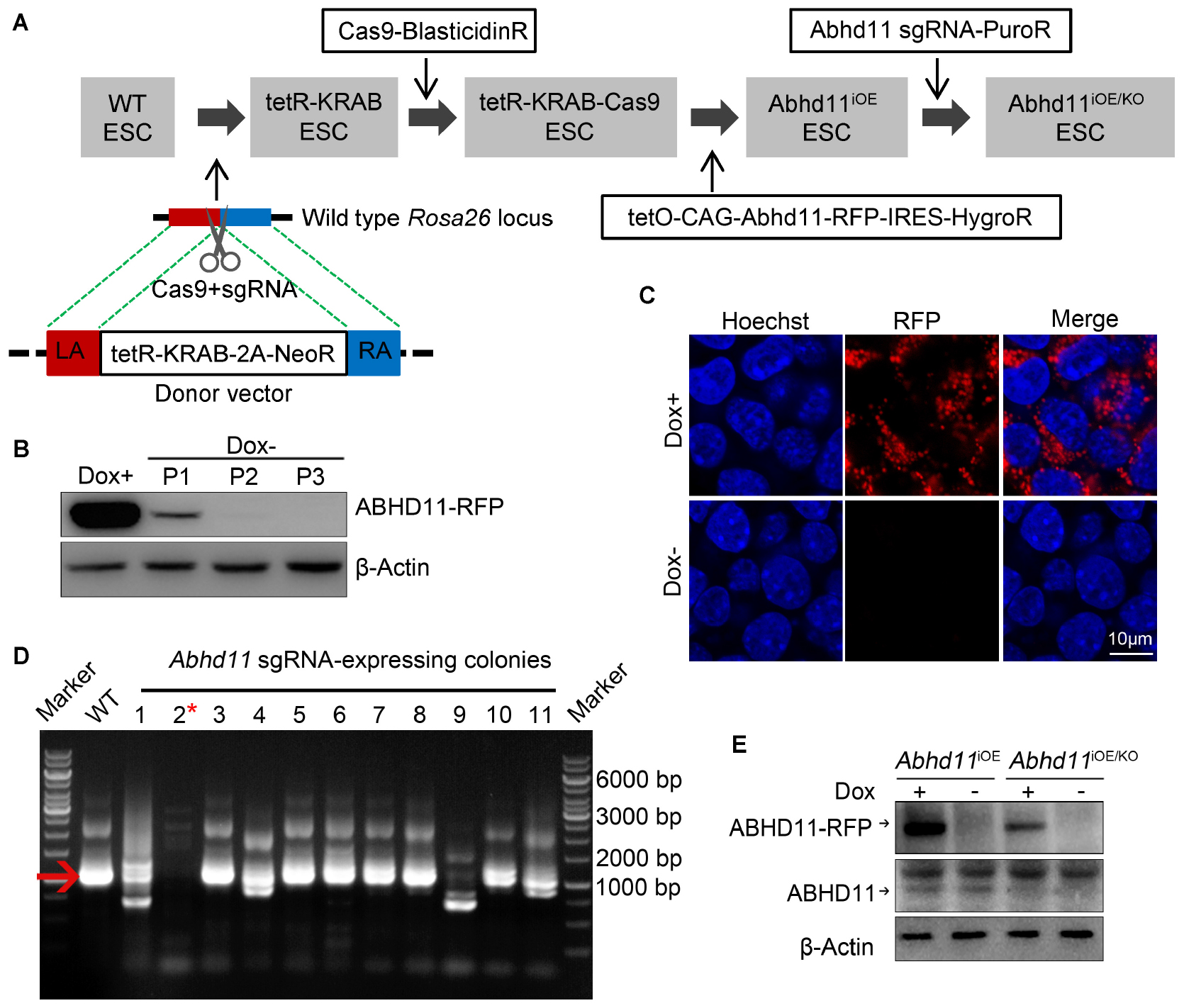
Figure 1. Generation and characterization of Abhd11iOE ESCs and Abhd11iOE/KO ESCs. (A) Schematic illustration of the four-step strategy for generating Abhd11iOE ESCs and Abhd11iOE/KO ESCs. Firstly, the tetR-Krab cassette was inserted into Rosa26 locus. Secondly, lentivirus expressing Cas9 was transduced into tetR-KRAB-expressing ESCs followed by selection with blasticidin (2 μg/ml). Thirdly, the cells were transduced with the expression cassette of tetO-CAG-Abhd11-RFP-IRES-HygroR and selected with Hygromycin (200 μg/ml) and Dox (100 ng/ml). Finally, the Abhd11iOE ESCs were infected with lentivirus expressing sgRNA against Abhd11 and selected using puromycin (1 μg/ml). LA, left arm; RA, right arm. (B) Western blot analysis of the RFP expression in Abhd11iOE ESCs cultured with Dox (1 μg/ml) or without Dox. Cells were subjected to serial passage (P) every 2 days. β-Actin was used as a loading control. (C) Fluorescence images of Abhd11iOE ESCs cultured with Dox (1 μg/ml) or without Dox. After fixation, cells were counterstained with hoechst. (D) RT-PCR showing the expression of endogenous Abhd11 mRNA in wild-type (WT) ESCs and Abhd11 sgRNA-expressing colonies. The red arrow indicates the specific RT-PCR product (1002 bp) of the endogenous Abhd11 mRNA. The red asterisk refers to a colony in which the endogenous Abhd11 gene was knocked out. (E) Western blot analysis of ABHD11 using anti-ABHD11 antibody. Abhd11iOE ESCs were cultured in the presence of 1 μg/ml Dox or without Dox. Abhd11iOE/KO ESCs were cultured in the presence of 1 ng/ml Dox or without Dox. β-Actin was used as a loading control.
ABHD11 Contributes to Expansion of ESCs
To begin with, we characterized the phenotype of Abhd11-null ESCs. When plated in culture, Abhd11KO ESCs formed smaller colonies (Figure 2A), and concurrently displayed a marked reduction in cell numbers compared to control cells (Figure 2B). Moreover, colony formation assays showed that the number of colonies formed by Abhd11KO ESCs was less than the control cells (Figure 2C). Colony morphology, cell number, and colony-forming potential were restored in Abhd11RES ESCs, and were indistinguishable from those of control cells (Figures 2A–C), indicating that the observed phenotypes in Abhd11KO ESCs were specifically due to loss of ABHD11 function, rather than to possible off-target effects of CRISPR/Cas9 (Fu et al., 2013). However, in contrast to ESCs, MEFs expanded normally when infected with lentivirus expressing Cas9-sgRNA targeting Abhd11 (Supplementary Figure S3), which suggested that the role of ABHD11 in the expansion of ESCs was cell context dependent. In contrast to Abhd11KO ESCs, Abhd11OE ESCs exhibited faster growth compared to RFP OE (RFPOE) ESCs (Figures 2D–F), indicating that Abhd11 OE promoted ESC expansion.
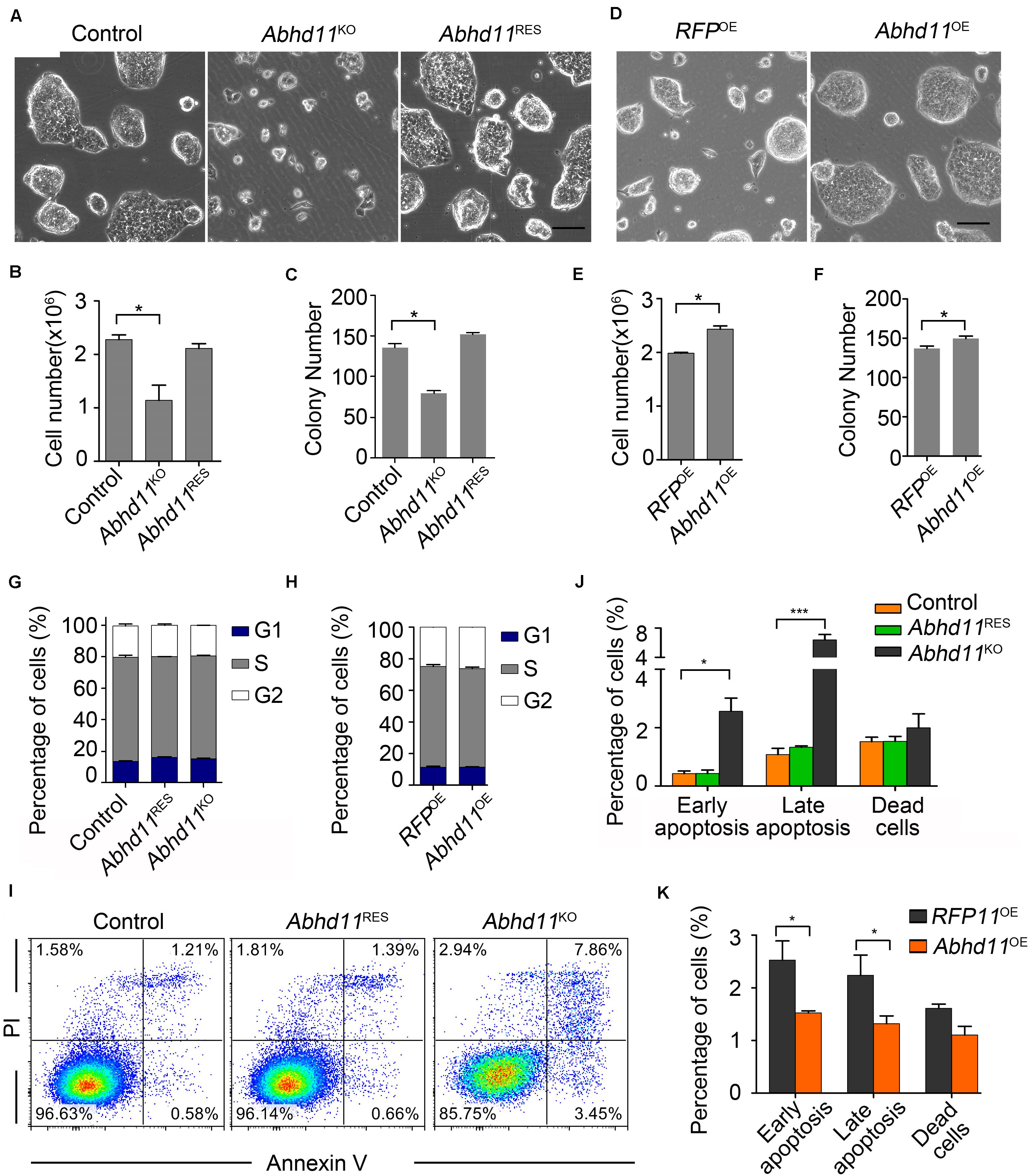
Figure 2. ABHD11 contributes to the expansion of ESCs. (A) Representative morphology of colonies formed by control, Abhd11KO, and Abhd11RES ESCs. Scale bar, 100 μm. (B) Cell growth of control, Abhd11KO, and Abhd11RES ESCs. (C) Quantitative analysis of colonies formed by control, Abhd11KO and Abhd11RES ESCs. (D) Representative morphology of colonies formed by the indicated cells. Scale bar, 100 μm. (E) Cell growth of RFPOE ESCs and Abhd11OE ESCs. (F) Quantitative analysis of colonies formed by RFPOE and Abhd11OE ESCs. (B,E) Cells (1 × 105 cells/well in 12-well plates) were cultured for 2 days and cell numbers were counted. (C,F) Cells were plated at clonal density (200 cells/well in 12-well plates) and cultured for 5 days. (G,H) Proportions of the cells in G1, S and G2 phase. Cells were stained with propidium iodide (PI) and analyzed by fluorescence-activated cell sorting (FACS). (I) Apoptosis of the indicated cells was measured by FACS with Annexin-V and PI staining. One of three experiments with similar results was shown. (J,K) The fraction of the indicated cells labeled for early apoptosis (Annexin V+PI–), late apoptosis (Annexin V+PI+), and dead cells (Annexin V–PI+). Data in panels (B,C,E–H,J,K) were represented as mean ± s.d.; n = 3. *p < 0.05, ***p < 0.001.
Opposite Effects of Abhd11 KO and OE on Apoptosis
The opposite effects of Abhd11 KO and OE on cell expansion led us to explore whether ABHD11 contributed to the regulation of the cell cycle or apoptosis. Cell cycle analysis showed that both Abhd11KO and Abhd11OE ESCs showed a normal cell cycle distribution (Figures 2G,H). However, we found that the number of apoptotic cells, as determined by Annexin V, significantly increased in Abhd11KO ESCs (Figures 2I, 4J), suggesting that the loss of ABHD11 triggered apoptosis. In contrast, the fraction of apoptotic and necrotic cells among Abhd11OE ESCs was approximately 50% of that of RFPOE ESCs under different culture conditions, including maintenance, serum starvation, differentiation in the absence of leukemia inhibitory factor (LIF) or addition of retinoic acid (Figure 2K, and Supplementary Figure S4). These data showed that Abhd11 OE exerted an anti-apoptotic effect in ESCs.
Abhd11 KO Induces Autophagy
It has been reported that autophagy (literally, ‘self-eating’), which is responsible for the maintenance of cellular homeostasis under metabolic stress conditions, plays a key role in the regulation of cell survival and death (Klionsky et al., 2016). We thus tried to address whether Abhd11 KO in ESCs could induce autophagy. We found an increase in the amount of the active form of microtubule-associated protein light chain 3 (LC3-II), a marker of autophagy, in Abhd11KO ESCs, compared to control ESCs (Figure 3A). We then transiently transfected cells with green fluorescent protein (GFP)-fused LC3 (GFP-LC3) to monitor the autophagic activity (Klionsky et al., 2016). Immunofluorescence confocal microscopy showed that GFP-LC3 was found to distribute evenly throughout the cytoplasm with few punctate spots in both control and Abhd11RES cells. However, large typical GFP-LC3 dots, indicative of cytoplasmic autophagosomes, were observed in Abhd11KO ESCs (Figures 3B,C). Transmission electron microscopy (TEM) detection also verified that large double-membrane autophagic vacuoles were present in Abhd11KO ESCs (Figure 3D). In addition, when cultured in the presence chloroquine, which is a lysosomal inhibitor and prevents the degradation of LC3-II, both LC3II and large typical GFP-LC3 dots were increased in Abhd11KO ESCs, compared to control cells (Supplementary Figure S5). Collectively, these results demonstrated that Abhd11 depletion induced autophagy in ESCs.
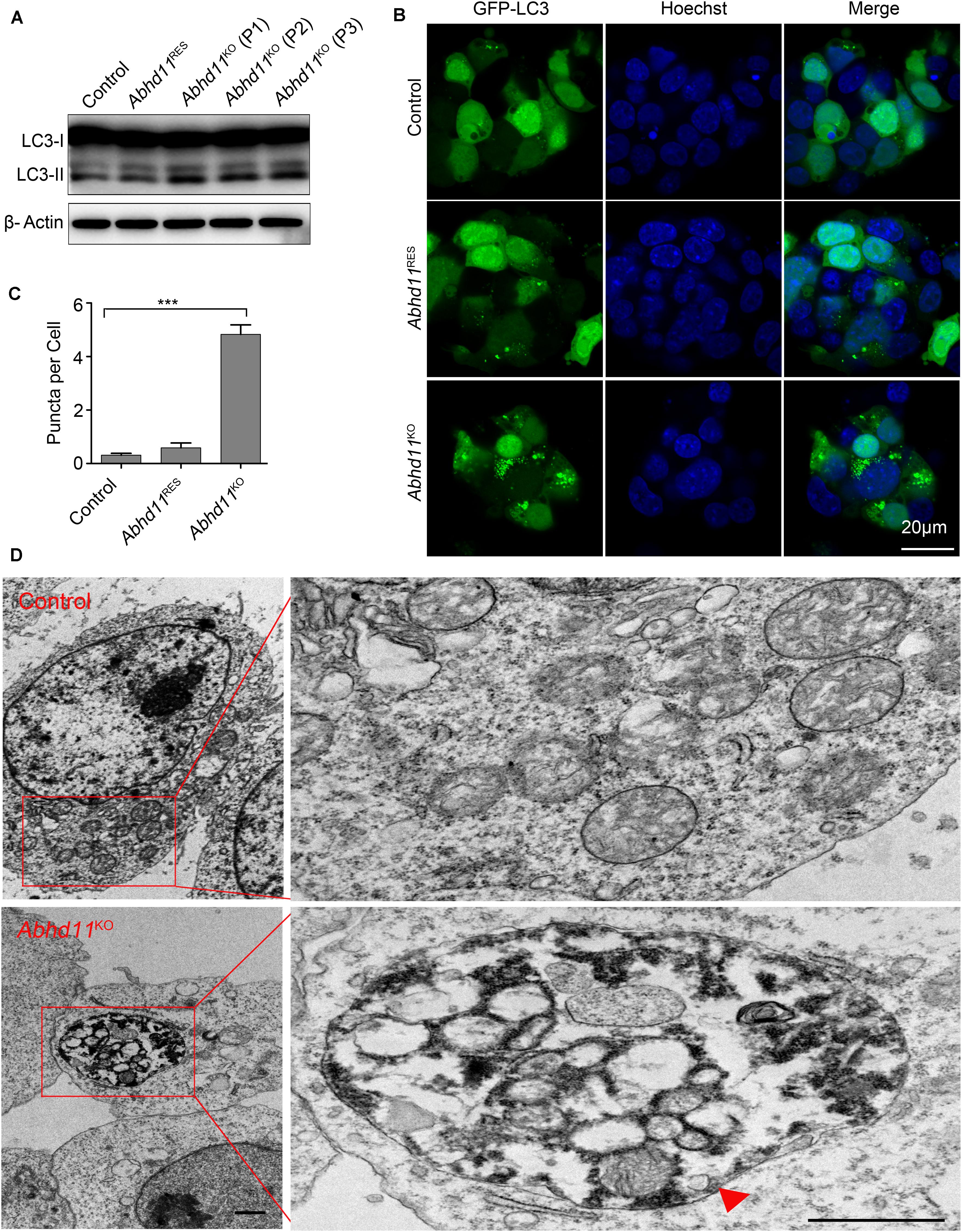
Figure 3. Abhd11 KO induces autophagy. (A) Western blot analysis of LC3-I and LC3-II expression level in the indicated cells during passage (P) with anti- LC3 antibody. (B) Confocal fluorescence microscopy images showing the subcellular localization of GFP-LC3 in the indicated cells. (C) Quantification of GFP-LC3 puncta in the indicated cells. Data were represented as mean ± s.d. from three independent experiments and at least 50 cells in each experiment were counted. ***p < 0.001. (D) Representative TEM images of control ESCs and Abhd11KO ESCs. A big autophagic vacuole appeared as double-membrane structure was highlighted with a red triangle. Scale bar, 1 μm.
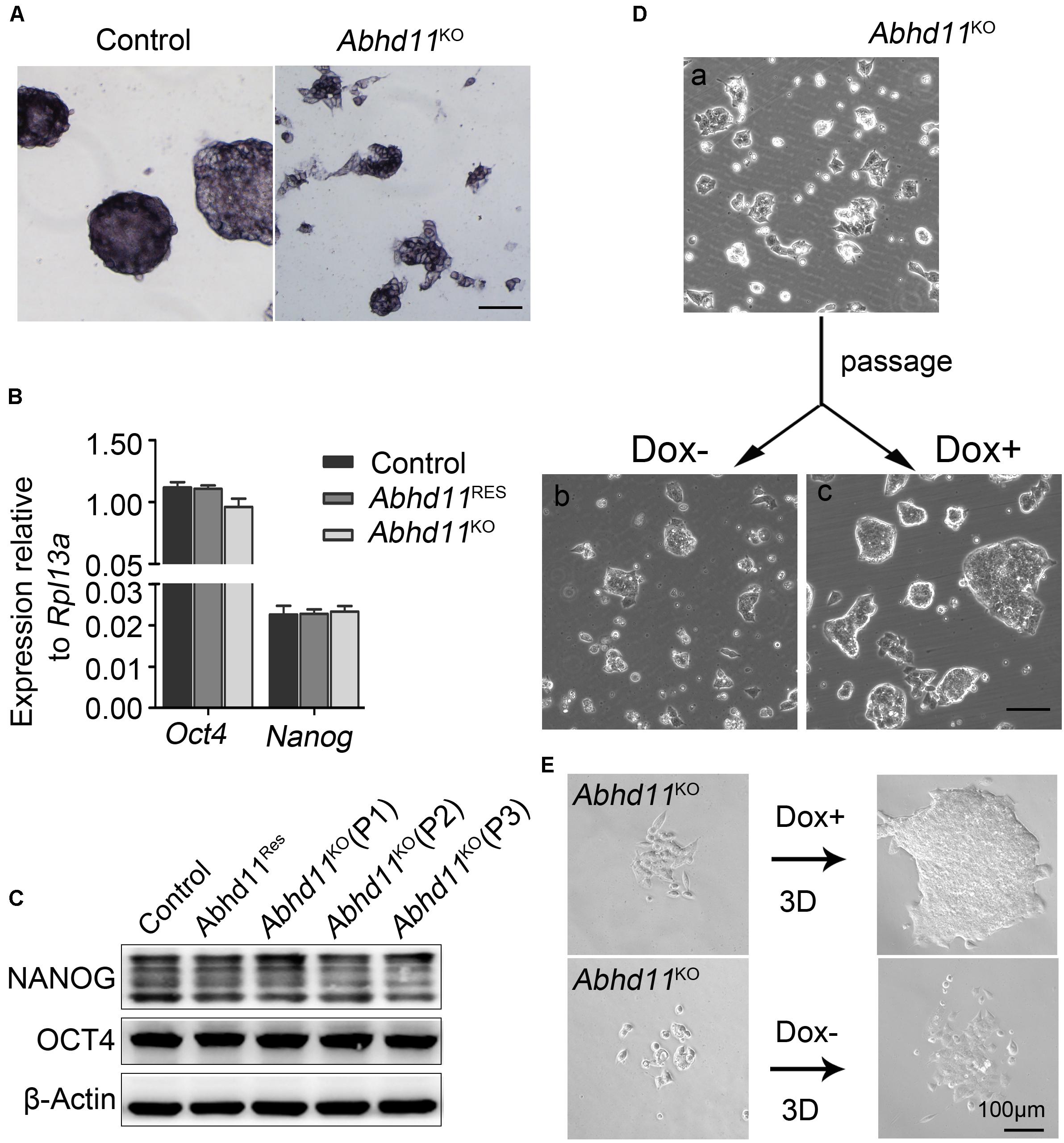
Figure 4. Abhd11 is dispensable for the maintenance of pluripotency. (A) Alkaline phosphatase staining of colonies formed by the indicated cells. Scale bar, 100 μm. (B) qRT-PCR analysis of Oct4 and Nanog mRNA level in the indicated cells. (C) Western blot analysis of OCT4 and NANOG expression level in the indicated cells during passage. (D) Representative morphology of colonies formed by Abhd11KO ESCs at high colony density. (E) Morphologies of colonies formed by Abhd11KO ESCs at low colony density.
ABHD11 Is Dispensable for the Maintenance of Pluripotency
Next, we attempted to address whether Abhd11 KO triggered ESCs to exit pluripotency and enter differentiation. We observed that the colonies formed by Abhd11KO ESCs were still alkaline phosphatase-positive (Figure 4A), which was indicative of undifferentiated ESCs. In addition, Abhd11KO ESCs displayed no changes in the expression of the pluripotency markers Oct4 and Nanog (Figures 4B,C). Notably, Abhd11KO ESCs could be maintained during serial passages (P) and rapidly recovered the ability to expansion and formed typical compact colonies at both high and low cell density when Dox was re-added to the medium (Figures 4D,E). These data indicated that Abhd11KO ESCs maintained a pluripotent state without commitment to differentiation. In addition, we observed that Abhd11OE ESCs also displayed no changes in alkaline phosphatase staining and Oct4 protein level compared to RFPOE ESCs (Supplementary Figures S6A,B).
ABHD11 Is Necessary for the Full Execution of Differentiation Programs
When plated on gelatin at clonal density and induced to differentiation in the absence of LIF, Abhd11KO ESCs appeared smaller compared to the control cells (Figure 5A). In addition, the embryoid bodies (EBs) generated by Abhd11KO ESCs were smaller and fewer than in control cells (Figures 5B,C). Furthermore, while both Abhd11KO and control ESCs formed teratomas in vivo, the average size of teratomas formed by Abhd11KO ESCs was significantly smaller compared to control ESCs (Figures 5D,E). To assess whether Abhd11 deficiency affected the differentiation ability of ESCs in vitro, the expression patterns of pluripotency-associated genes and lineage-specific gene markers were subsequently assessed by quantitative reverse transcriptase-PCR (qRT-PCR). When induced to differentiate, Abhd11-KO cells failed to fully downregulate Oct4 and Nanog (Figure 5F) upon cellular differentiation (Pesce and Scholer, 2001; Chambers et al., 2003). Moreover, under these conditions, Abhd11-KO cells did not normally induce the expression of early differentiation markers, such as Gata6 (Endoderm), Sox1 (Ectoderm), and Flk1 (Mesoderm) (Figure 5F). In addition, although teratomas derived from Abhd11KO ESCs contained cell types representing all three germ layers, Abhd11KO ESCs derived little cells with vacuoles while control cells formed endodermal monolayer columnar epithelium consisting of enriched cells with vacuoles (Figure 5G). Taken together, these data demonstrated that ABHD11 was necessary for the full execution of differentiation programs. Moreover, ABHD11 OE could not block the differentiation of ESCs in the absence of LIF and in vivo (Supplementary Figures S6C–E).
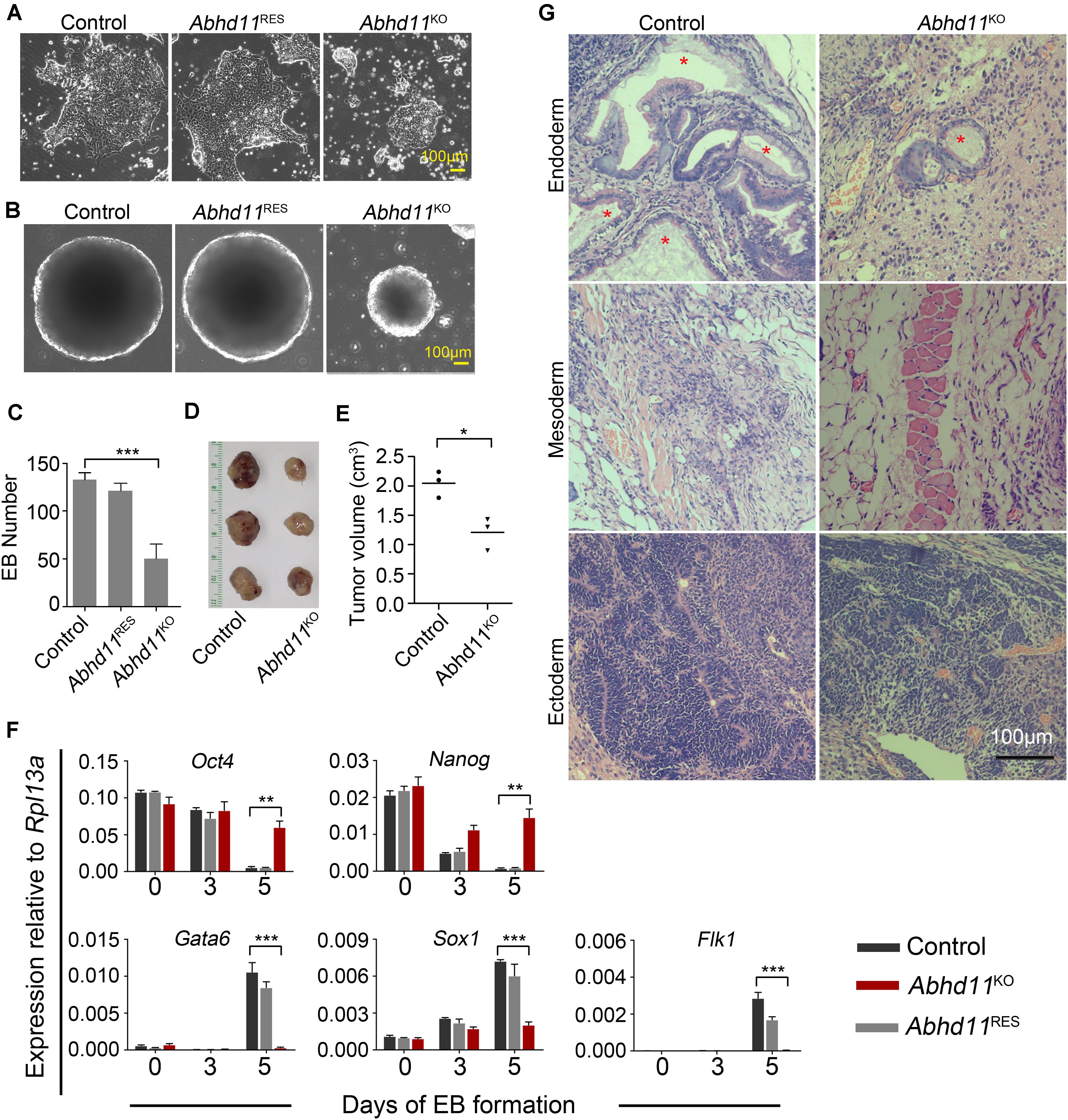
Figure 5. ABHD11 is necessary for the full execution of differentiation programs. (A) Representative images of the indicated cells cultured for 4 days in the absence of LIF. (B) Representative images of EBs formed by the indicated cells at day 5. (C) Quantification of EBs formed by the indicated cells at day 5. (D) Teratomas formed from the control and Abhd11KO ESCs. Teratomas were collected 4 weeks after the injection of ESCs subcutaneously into nude mice. (E) The volume of teratomas presented in panel (D). (F) qRT-PCR analysis of expression levels of pluripotency and germ layer marker during the course of EB formation. The EBs were collected at Day 0, 3, 5 during the course of EB formation. All data were normalized to Rpl13a. (G) Teratoma section were stained with hematoxylin/eosin. Asterisks indicate typical endodermal epithelium–like structures (simple columnar epithelium). Data in panels (C,F) were represented as means ± s.d.; n = 3. *p < 0.05, **p < 0.01, ***p < 0.001.
Abhd11 KO Results in Disturbances in Signal Transduction
Recently, lipid metabolic intermediates have been demonstrated to participate in cellular signaling processes (Zechner et al., 2012). In light of the predicted role of ABHD11 in lipid metabolism (Arya et al., 2017), we then tried to identify which signaling pathways in ESCs could be affected by Abhd11 depletion. It is well-known that ESCs can be maintained under defined culture conditions with the addition of LIF, which induces the activation of signal transducer and activator of transcription 3 (Stat3) and AKT (also known as protein kinase B, PKB) signaling (Niwa et al., 2009). We found that phosphorylated Stat3, total Stat3, and total AKT protein levels were not changed in Abhd11KO ESCs (Figure 6A). However, there was an increase in AKT phosphorylation at Ser473, and a reduction of AKT phosphorylation at Thr308 in P3 Abhd11KO ESCs (Figure 6A). This suggested a dysregulation of AKT activity in these cells. Nevertheless, the total level of AKT activity is difficult to assess, as it depends on the extent of phosphorylation at both Thr308 and Ser473 residues, as well as on its total protein concentration (Alessi et al., 1996).
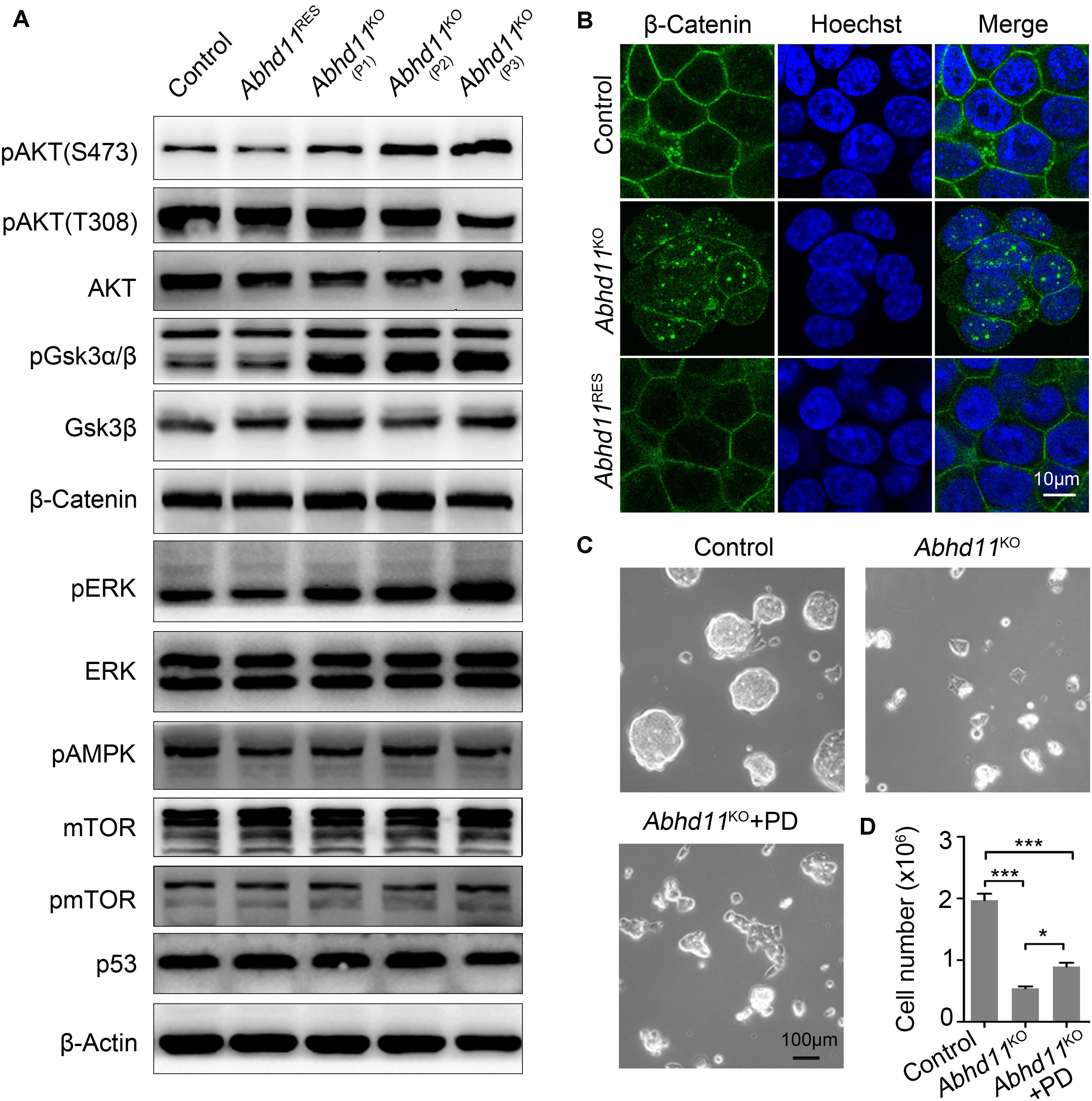
Figure 6. Abhd11 KO results in disturbances in signal transduction. (A) Western blot analysis using antibodies as indicated. (B) Immunofluorescence analysis of β-Catenin in the indicated cells. Cells were counterstained with hoechst. (C) Representative morphology of colonies formed by control and P3 Abhd11KO ESCs. Abhd11KO ESCs were plated in duplicate and treated with 1 μM PD0325901 (Abhd11KO + PD). (D) Cell numbers in panel C were counted. Data were represented as mean ± s.d. from three independent experiments. *p < 0.05, ***p < 0.001.
Next, we found that, after Dox withdrawal, the extent of phosphorylation of glycogen synthase kinase 3 beta (GSK3β) (at Serine-9), was significantly increased in Abhd11KO ESCs (Figure 6A). This suggested that GSK3β signaling was sensitive to the loss of ABHD11. It has been reported that Serine-9-phosphorylated GSK3β inhibits enzyme activity and leads to the nuclear accumulation of β-Catenin (Sato et al., 2004; Ding et al., 2005). Consistently, immunofluorescent staining showed an increase in β-Catenin translocation from the cytoplasm to the nucleus in Abhd11KO ESCs (Figure 6B). These data indicated that Abhd11 depletion had an impact on the regulation of the GSK3β/β-Catenin signaling pathway.
We also found that the extent of phosphorylation of the extracellular signal-regulated kinase (ERK) increased in Abhd11KO ESCs (Figure 6A). Inhibition of ERK signaling enhances the self-renewal activity of ESCs (Burdon et al., 1999). To verify whether a higher extent of ERK phosphorylation could account for defective self-renewal in Abhd11KO ESCs, we treated Abhd11KO ESCs with the specific and widely used ERK phosphorylation inhibitor, PD0325901. We found that treatment with PD0325901 could not completely reverse the Abhd11 KO-induced phenotype (Figures 6C,D), suggesting that additional factors were involved in the impairment of self-renewal observed in Abhd11KO ESCs. The protein level of p53 and mTOR, as well as the phosphorylation of mTOR and pAMPK, were not altered in Abhd11-KO cells (Figure 6A). In addition, the detected signalings in Abhd11OE ESCs showed no obvious changes (Supplementary Figure S6B).
Abhd11 KO Causes the Misexpression of Metabolic Genes
To gain insights into the molecular mechanisms underlying ABHD11-dependent maintenance of self-renewal in ESCs, we characterized the transcriptome of Abhd11KO ESCs by RNA seq. Notably, the Volcano Plot showed that the most significantly upregulated genes in Abhd11KO ESCs, encoded for enzymes mainly involved in amino acid metabolism, such as Aars, Nars, Sars, Cars, and Chac1 (Figure 7A). The most significant downregulated genes in Abhd11KO ESCs included those for rate-limiting glycolytic enzymes (pfkl, pkm), lipid synthesis enzymes (Scd1, Scd2, Fasn, Acly), and Lin28, which plays a cooperative role in the regulation of pluripotency and metabolic proteome (Zhang J. et al., 2016; Figure 7A). Further analysis showed that Abhd11 deletion caused a prominent upregulation of genes for amino acid activation, translocation, synthesis, fatty acid oxidation, and tricarboxylic acid cycle, and a substantial downregulation of key enzymes involved in lipid synthesis and of almost all glycolytic enzymes (Figures 7B,C, and Supplementary Table S2). Consistent with the above description, the expression of most of the pluripotency markers, such as Oct4, Nanog, Sox2, Dax1, Klf4, and so on, were not changed in Abhd11KO ESCs (Supplementary Table S2).
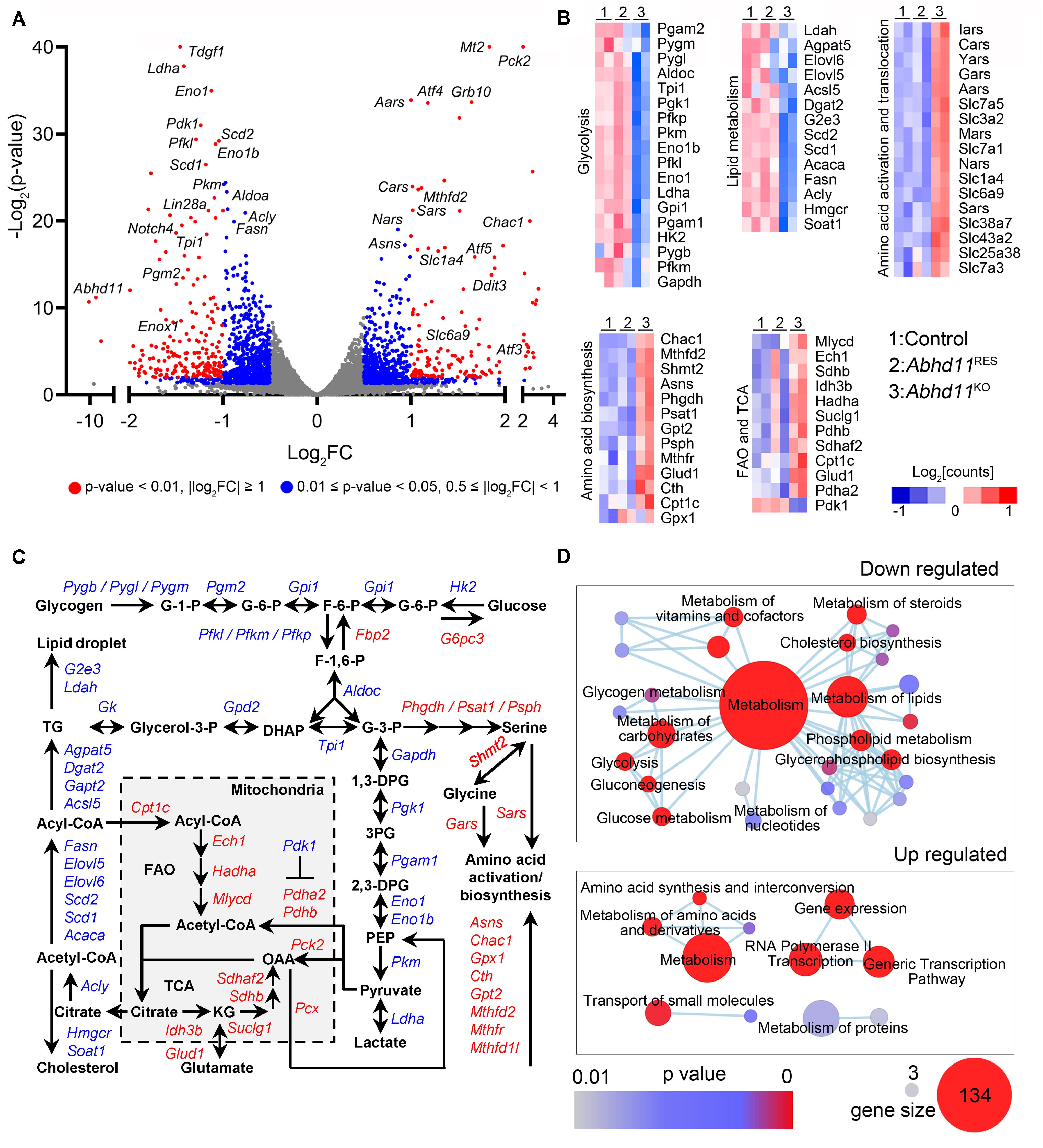
Figure 7. Abhd11 KO causes the misexpression of metabolic genes. (A) Volcano plot of DEGs in P3 Abhd11KO ESCs versus control ESCs. FC, fold change. (B) Heatmap of metabolic genes differentially expressed in Abhd11KO ESCs versus control ESCs. (C) Biochemical pathways of genes that are up-regulated (red) and down-regulated (blue) as a result of Abhd11 depletion. G-1-P, glucose 1-phosphate; G-6-P, glucose 6-phosphate; F-6-P, fructose 6-phosphate; F-1,6-P, fructose 1,6-biphosphate; G-3-P, glyceraldehyde 3-phosphate; DHAP, dihydroxy acetone phosphate; 1,3 DPG, 1,3-diphosphoglyceric acid; 3PG, 3-Phosphoglycerate; 2,3-DPG, 2,3-diphosphoglycerate; PEP, Phosphoenolpyruvate; OAA, oxaloacetate; FAO, fatty acid oxidation; TCA, citric acid cycle; KG, α-ketoglutarate; TG, triglyceride. (D) Enrichment map networks of reactome terms corresponding to down-regulated genes (upper panel, a part of the total networks that was presented in Supplementary Figure S7C) and up-regulated genes (lower panel) in P3 Abhd11KO ESCs versus control ESCs. Reactome terms were visualized by the cytoscape plug-in: Enrichment Map. Node size represents the gene-set size and node color represents enrichment significance (as indicated). The deposited data can be found at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142067.
Next, reactome in terms of the DEGs were analyzed and visualized by using Enrichment Map, an established Cytoscape plug-in (Reimand et al., 2019). In Abhd11KO ESCs, the downregulated genes were functionally enriched in lipid and carbohydrate metabolism, as well as signal transduction, while the upregulated genes were enriched in amino acid metabolism and regulation of gene expression (Figure 7D and Supplementary Figure S7A). Consistently, gene ontology (GO) analyses of the DEGs, also revealed an enrichment in metabolic processes (Supplementary Table S3). Collectively, these data suggested that the loss of ABHD11 resulted in dysregulated expression of genes involved in multiple metabolic events.
Furthermore, we also analyzed DEGs between Abhd11OE and control ESCs, and identified 261 downregulated and 216 upregulated genes in Abhd11OE ESCs (Supplementary Figure S7B and Supplementary Table S2). The total number of deregulated genes in Abhd11OE ESCs was much lower than in Abhd11KO ESCs, suggesting a lower impact of Abhd11 OE on overall transcription, compared to Abhd11 KO. Notably, there was little overlap between DEGs in Abhd11OE ESCs and DEGs in Abhd11KO ESCs (Supplementary Figure S7C). In addition, the DEGs in Abhd11OE ESCs were not enriched in metabolic genes and pluripotency related genes (Supplementary Table S2). These data suggested that Abhd11 OE exerted little effects on the expression of metabolic genes.
ABHD11 Contributes to the Homeostasis of Lipid Metabolism
To address the role of ABHD11 in lipid metabolism, we firstly detected the protein level of FASN (Fatty Acid Synthase), which is a key enzyme in de novo lipogenesis. Consistent with the mRNA expression, the result showed that the expression of FASN was downregulated in Abhd11KO ESCs (Figure 8A). The downregulation of key enzymes in lipid synthesis in Abhd11KO ESCs suggested inhibition of de novo lipogenesis. Then we analyzed the lipid profiles by LC-MS/MS. Principal component analysis (PCA) of lipid metabolites indicated clear differences among control, Abhd11KO, and Abhd11OE ESCs (Figure 8B). Among the 30 lipid classes detected in ESCs, digalactosyldiacylglycerol and sulfoquinovosyldiacylglycerol (saccharolipids), triglyceride and diglyceride (glycerolipids), phosphatidylethanolamine and phosphatidylglycerol (glycerophospholipids) content showed a significant increase in Abhd11KO ESCs while a significant decrease in Abhd11OE ESCs (Figures 8C,D and Supplementary Table S6). In addition, KO of Abhd11 increased the levels of fatty acid and acyl carnitine (fatty acyls) while reduced the levels of ceramides and sphingosine (sphingolipids) (Figures 8C,D and Supplementary Table S6). Volcano plots showed that the lipid species showing the most significant change in content were triglyceride and ceramides in both Abhd11KO and Abhd11OE ESCs (Figures 8E,F and Supplementary Table S7). The triglyceride content decreased in Abhd11OE ESCs while increased in Abhd11KO ESCs suggesting that ABHD11 appeared to have substrate preference of triglyceride. Finally, we observed lipid droplets (LD) formation and fusion as well as LD-containing autophagosome (lipophagosome) in Abhd11KO ESCs (Figure 8G), suggesting that Abhd11 KO induced LD growth and lipophagy. Taken together, ABHD11 functions as a key regulator of lipid metabolism in ESCs.
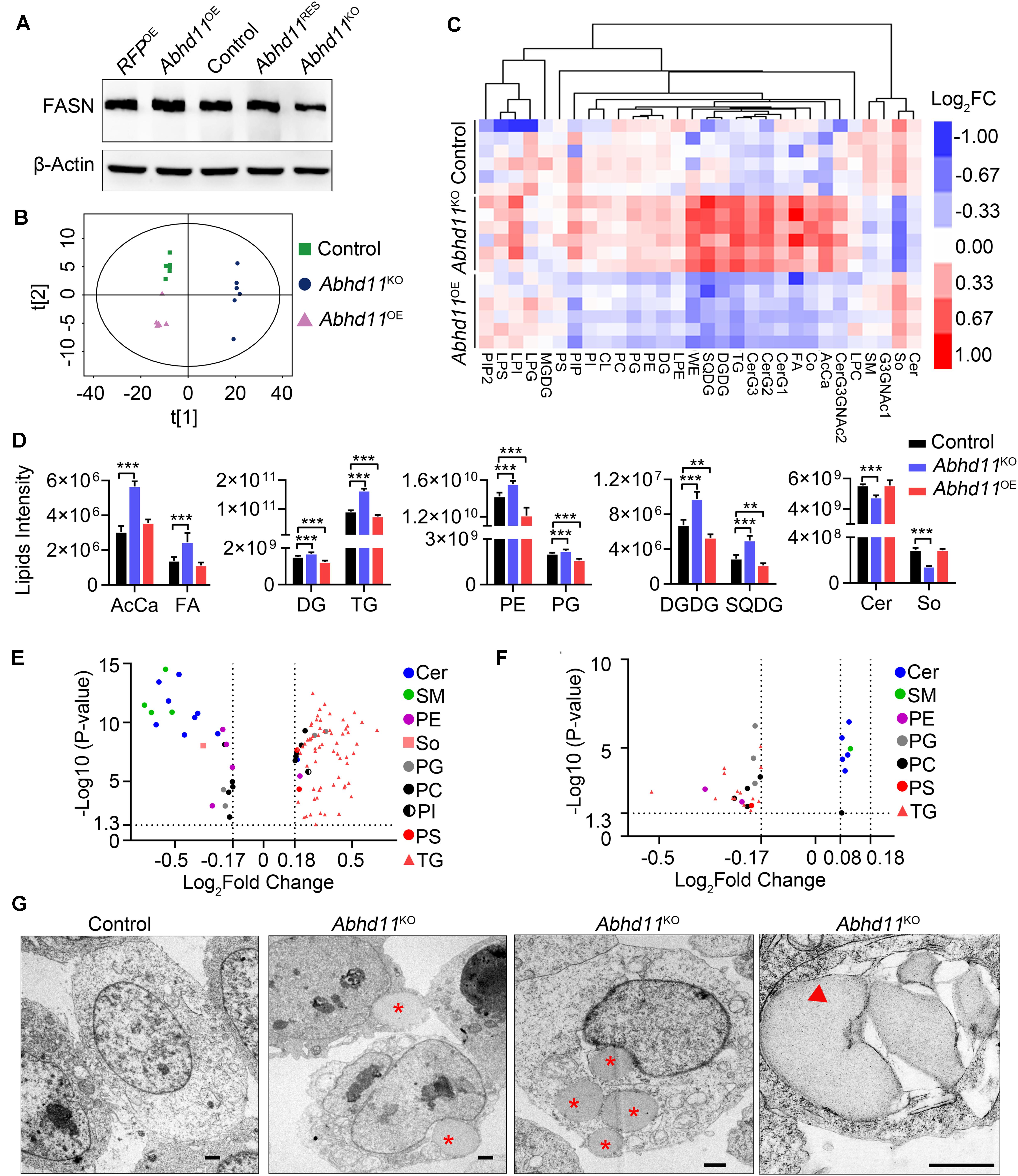
Figure 8. ABHD11 contributes to the homeostasis of lipid metabolism. (A) Western blot analysis of FASN expression level in the indicated cells. (B) Principal component analysis of lipid metabolites in control, Abhd11KO, and Abhd11OE ESCs. (C) Heatmap of the 30 lipid classes content in control, Abhd11KO, and Abhd11OE ESCs. AcCa, Acyl Carnitine; Cer, Ceramides; CL, Cardiolipin; Co, Coenzyme; DG, diglyceride; DGDG, Digalactosyldiacylglycerol; FA, Fatty acid; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; LPG, lysophosphatidylglycerol; LPI, lysophosphatidylinositol; MGDG, Monogalactosyldiacylglycerol; So, Sphingosine; SM, sphingomyelin; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PIP, phosphatidylinositol; PS, phosphatidylserine; TG, triglyceride; SQDG, Sulfoquinovosyldiacylglycerol; WE, wax exters. (D) Lipid intensity of lipid classes altered in Abhd11KO and Abhd11OE ESCs measured by LC-MS/MS. Data were represented as mean ± s.d. from six biologically independent samples. **p < 0.01, ***p < 0.001. (E) Volcano plot of lipid species that showed the most significant change in Abhd11KO ESCs compared to control (fold change (FC) > 1.5 or FC < 0.67, P-value < 0.05, VIP > 1). (F) Volcano plot of lipid species that showed the most significant change in Abhd11OE ESCs compared to control (FC > 1.2 or FC < 0.67, P-value < 0.05, VIP > 1). (G) Representative TEM images of LD (indicated by a red asterisk) and lipophagosome, which appeared as double-membrane structure and contained LDs (indicated by a red triangle), in Abhd11KO ESCs. Scale bar, 1 μm.
Discussion
ABHD11 was previously identified as a conserved lipase across species and has a pivotal role in lipid metabolism in Arabidopsis and yeast (Vijayakumar et al., 2016; Arya et al., 2017). In human, ABHD11 is one of several genes deleted in Williams–Beuren syndrome, a developmental disorder associated with haploinsufficiency of multiple genes (Merla et al., 2002). In addition, ABHD11 is a potential biomarker of lung adenocarcinoma (Wiedl et al., 2011), and is linked to breast cancer malignancy (Heinonen et al., 2015). Despite such reports, however, the actual physiological functions of ABHD11 are unclear. In this study, we characterized the function of ABHD11 in the maintenance of ESCs by both loss- and gain-of-function experiments.
Although ABHD11 has a conserved lipase motif, its endogenous substrates and products are still unknown. In Arabidopsis, the abolishment of ABHD11 activity increases the level of polar lipids in the leaves (Vijayakumar et al., 2016). In yeast, the overexpression of human ABHD11 or its yeast homolog causes a reduction in the levels of non-polar lipids (Arya et al., 2017). In this study, we observed the opposite effects of OE and KO of ABHD11 on the lipid classes content, such as glycerolipids and glycerophospholipids, supporting its role as a lipid hydrolase. Notably, Abhd11 KO resulted in the downregulation of enzymes for lipid synthesis, and reduced sphingolipids content in mouse ESCs. It is possible that hydrolyses of substrates by ABHD11 may affect signaling processes contributing to de novo lipogenesis in ESCs. In addition, as sphingolipids function as regulators of crosstalk between apoptosis and autophagy (Young et al., 2013), sphingolipids mediates the function of ABHD11 in the expansion and survival of ESCs is an interesting possibility that is worthy of further investigation.
A recent study revealed that regulation of de novo lipid synthesis by the lipogenic enzyme, acetyl-CoA carboxylase alpha (ACACA), is critical for ESC pluripotency (Wang et al., 2017). The study also demonstrated that ACACA overexpression and knockdown lead to increased and decreased expression, respectively, of pluripotent genes such as Oct4 and Sox2 (Wang et al., 2017). However, we observed that Abhd11KO ESCs exhibited decreased lipid synthesis and self-renewal ability, while maintaining a pluripotent state, as well as normal expression of Oct4 and Nanog, suggesting that ABHD11 preserved ESC identity by a mechanism distinct from that of ACACA.
When Abhd11 expression was suppressed in ESCs, the level of GSK3β phosphorylation significantly increased, indicating a role of ABHD11 in the regulation of GSK3β signaling pathways. The GSK3β is a multifunctional serine/threonine kinase involved in a wide range of cellular processes, ranging from glycogen metabolism to cell cycle regulation and proliferation (Doble and Woodgett, 2003). In ESCs, the inhibition of GSK3β promotes self-renewal through the activation of β-Catenin signaling (Sato et al., 2004; Ying et al., 2008). Thus, enhanced β-Catenin signaling pathway may not be the cause of impaired self-renewal ability in Abhd11-depleted ESCs. Notably, RNAseq showed that the expressions of most of the pluripotency regulators were not changed in Abhd11KO ESCs (Supplementary Table S2). It is possible that the elevated levels of phosphorylation of GSK3β and nuclear accumulation of β-Catenin were insufficient to increase the expression of the pluripotency genes. Nevertheless, GSK3β inhibition can induce autophagy and cell death (Yang et al., 2010), while GSK3β activation was reported to be either anti-apoptotic or pro-apoptotic (Beurel and Jope, 2006), indicating an impact of GSK3β activity on autophagy and apoptosis. Thus, whether GSK3β inhibition caused by Abhd11 KO contributes to autophagy or apoptosis is still an open issue.
Notably, our study showed that Abhd11 overexpression and deletion increased and decreased the expansion of ESCs, respectively, and that Abhd11 suppression downregulated the expression of metabolic genes related to both lipid biosynthesis and glycolysis. In consideration of the high expression of ABHD11 in lung cancer (Wiedl et al., 2011; Heinonen et al., 2015) and the key roles of both elevated de novo lipid biosynthesis and increased glycolysis in cancer (DeBerardinis and Chandel, 2016), it is crucial to establish whether ABHD11 contributes to malignancy and whether its inhibition can block tumor growth.
In summary, to our knowledge, this study is the first to provide evidence that ABHD11 functions as a key determinant of self-renewal and metabolic homeostasis in ESCs. Our novel discovery explored the broad role of ABHD proteins in lipid metabolism and stem cell biology and has offered new insights into the metabolic regulation of stem cell activity. However, the detailed mechanisms of the suppression of Abhd11 gene leading to the misexpression of metabolic genes, activation of autophagy, and the disturbances in signal transduction have been left unclarified. Future studies will be required to molecularly dissect the crosstalk among the processes that result from the KO of ABHD11 in ESCs.
Data Availability Statement
The datasets generated for this study can be found in the GSE142067.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethical and Experimental Committee of the Third Military Medical University.
Author Contributions
GL and YR contributed to the conception and design, collection and assembly of the data, data analysis, interpretation, manuscript writing, and financial support. JZ, YT, and JW analyzed the data and interpreted the study, and contributed to the manuscript revision, and final approval of the manuscript. WW, PH, and JX contributed to the administrative support, provision of study material and suggestion. XW, YC, LL, and YY collected and assembled the data, analyzed the data, and interpreted the study. RJ contributed to the conception and design, financial support, data analysis and interpretation, manuscript writing, and final approval of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31601108, 31571442, and 81702294).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Ian Chambers for the pPyCAGIP plasmid, and Hong Zheng for the pGFP-LC3 plasmid. We thank Lan Xiao for her support with our research.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.00570/full#supplementary-material
Footnotes
References
Alessi, D. R., Andjelkovic, M., Caudwell, B., Cron, P., Morrice, N., Cohen, P., et al. (1996). Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 15, 6541–6551. doi: 10.1002/j.1460-2075.1996.tb01045.x
Arya, M., Srinivasan, M., and Rajasekharan, R. (2017). Human alpha beta hydrolase domain containing protein 11 and its yeast homolog are lipid hydrolases. Biochem. Biophys. Res. Commun. 487, 875–880. doi: 10.1016/j.bbrc.2017.04.145
Beurel, E., and Jope, R. S. (2006). The paradoxical pro- and anti-apoptotic actions of GSK3 in the intrinsic and extrinsic apoptosis signaling pathways. Prog. Neurobiol. 79, 173–189. doi: 10.1016/j.pneurobio.2006.07.006
Boyer, L. A., Lee, T. I., Cole, M. F., Johnstone, S. E., Levine, S. S., Zucker, J. P., et al. (2005). Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956. doi: 10.1016/j.cell.2005.08.020
Burdon, T., Stracey, C., Chambers, I., Nichols, J., and Smith, A. (1999). Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev. Biol. 210, 30–43. doi: 10.1006/dbio.1999.9265
Chambers, I., Colby, D., Robertson, M., Nichols, J., Lee, S., Tweedie, S., et al. (2003). Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643–655. doi: 10.1016/s0092-8674(03)00392-1
Chu, V. T., Weber, T., Graf, R., Sommermann, T., Petsch, K., Sack, U., et al. (2016). Efficient generation of Rosa26 knock-in mice using CRISPR/Cas9 in C57BL/6 zygotes. BMC Biotechnol. 16:4. doi: 10.1186/s12896-016-0234-4
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Cornacchia, D., Zhang, C., Zimmer, B., Chung, S. Y., Fan, Y., Soliman, M. A., et al. (2019). Lipid deprivation induces a stable, naive-to-primed intermediate state of pluripotency in human PSCs. Cell Stem Cell 25, 120.e10–136.e10. doi: 10.1016/j.stem.2019.05.001
DeBerardinis, R. J., and Chandel, N. S. (2016). Fundamentals of cancer metabolism. Sci. Adv. 2:e1600200. doi: 10.1126/sciadv.1600200
Ding, Q. Q., Xia, W. Y., Liu, J. C., Yang, J. Y., Lee, D. F., Xia, J. H., et al. (2005). Erk associates with and primes GSK-3 beta for its inactivation resulting in upregulation of beta-catenin. Mol. Cell. 19, 159–170. doi: 10.1016/j.molcel.2005.06.009
Doble, B. W., and Woodgett, J. R. (2003). GSK-3: tricks of the trade for a multi-tasking kinase. J. Cell Sci. 116(Pt 7), 1175–1186. doi: 10.1242/jcs.00384
Fu, Y., Foden, J. A., Khayter, C., Maeder, M. L., Reyon, D., Joung, J. K., et al. (2013). High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826. doi: 10.1038/nbt.2623
Gu, W., Gaeta, X., Sahakyan, A., Chan, A. B., Hong, C. S., Kim, R., et al. (2016). Glycolytic metabolism plays a functional role in regulating human pluripotent stem cell state. Cell Stem Cell 19, 476–490. doi: 10.1016/j.stem.2016.08.008
Hassani, S. N., Totonchi, M., Gourabi, H., Scholer, H. R., and Baharvand, H. (2014). Signaling roadmap modulating naive and primed pluripotency. Stem Cells Dev. 23, 193–208. doi: 10.1089/scd.2013.0368
Heinonen, H., Lepikhova, T., Sahu, B., Pehkonen, H., Pihlajamaa, P., Louhimo, R., et al. (2015). Identification of several potential chromatin binding sites of HOXB7 and its downstream target genes in breast cancer. Int. J. Cancer 137, 2374–2383. doi: 10.1002/ijc.29616
Kim, J. (2008). An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132, 1049–1061. doi: 10.1016/j.cell.2008.02.039
Klionsky, D. J., Abdelmohsen, K., Abe, A., Abedin, M. J., Abeliovich, H., Arozena, A. A., et al. (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222. doi: 10.1080/15548627.2015.1100356
Kondoh, H., Lleonart, M. E., Nakashima, Y., Yokode, M., Tanaka, M., Bernard, D., et al. (2007). A high glycolytic flux supports the proliferative potential of murine embryonic stem cells. Antioxid. Redox. Signal. 9, 293–299. doi: 10.1089/ars.2006.1467
Lass, A., Zimmermann, R., Haemmerle, G., Riederer, M., Schoiswohl, G., Schweiger, M., et al. (2006). Adipose triglyceridelipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 3, 309–319. doi: 10.1016/j.cmet.2006.03.005
Li, M., Liu, G. H., and Izpisua Belmonte, J. C. (2012). Navigating the epigenetic landscape of pluripotent stem cells. Nat. Rev. Mol. Cell Biol. 13, 524–535. doi: 10.1038/nrm3393
Loh, Y.-H., Wu, Q., Chew, J.-L., Vega, V. B., Zhang, W., Chen, X., et al. (2006). The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38, 431–440. doi: 10.1038/ng1760
Lord, C. C., Thomas, G., and Brown, J. M. (2013). Mammalian alpha beta hydrolase domain (ABHD) proteins: lipid metabolizing enzymes at the interface of cell signaling and energy metabolism. Biochim. Biophys. Acta 1831, 792–802. doi: 10.1016/j.bbalip.2013.01.002
M, N. K., V, B. S. C. T., G, K. V., B, C. S., Guntupalli, S., and J, S. B. (2016). Molecular characterization of human ABHD2 as TAG lipase and ester hydrolase. Biosci. Rep. 36:e00358. doi: 10.1042/BSR20160033
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. doi: 10.1126/science.1232033
Merla, G., Ucla, C., Guipponi, M., and Reymond, A. (2002). Identification of additional transcripts in the Williams-Beuren syndrome critical region. Hum. Genet. 110, 429–438. doi: 10.1007/s00439-002-0710-x
Moussaieff, A., Rouleau, M., Kitsberg, D., Cohen, M., Levy, G., Barasch, D., et al. (2015). Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell. Metab. 21, 392–402. doi: 10.1016/j.cmet.2015.02.002
Ng, H. H., and Surani, M. A. (2011). The transcriptional and signalling networks of pluripotency. Nat. Cell Biol. 13, 490–496. doi: 10.1038/ncb0511-490
Nichols, J., and Smith, A. (2011). The origin and identity of embryonic stem cells. Development 138, 3–8. doi: 10.1242/dev.050831
Niwa, H., Ogawa, K., Shimosato, D., and Adachi, K. (2009). A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 460, 118–122. doi: 10.1038/nature08113
Obinata, D., Takada, S., Takayama, K., Urano, T., Ito, A., Ashikari, D., et al. (2016). Abhydrolase domain containing 2, an androgen target gene, promotes prostate cancer cell proliferation and migration. Eur. J. Cancer 57, 39–49. doi: 10.1016/j.ejca.2016.01.002
Panopoulos, A. D., Yanes, O., Ruiz, S., Kida, Y. S., Diep, D., Tautenhahn, R., et al. (2012). The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 22, 168–177. doi: 10.1038/cr.2011.177
Peng, Y., Miao, H., Wu, S., Yang, W., Zhang, Y., Xie, G., et al. (2016). ABHD5 interacts with BECN1 to regulate autophagy and tumorigenesis of colon cancer independent of PNPLA2. Autophagy 12, 2167–2182. doi: 10.1080/15548627.2016.1217380
Pesce, M., and Scholer, H. R. (2001). Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells 19, 271–278. doi: 10.1634/stemcells.19-4-271
Reimand, J., Isserlin, R., Voisin, V., Kucera, M., Tannus-Lopes, C., Rostamianfar, A., et al. (2019). Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 14, 482–517. doi: 10.1038/s41596-018-0103-9
Ruan, Y., He, J., Wu, W., He, P., Tian, Y., Xiao, L., et al. (2017). Nac1 promotes self-renewal of embryonic stem cells through direct transcriptional regulation of c-Myc. Oncotarget 8, 47607–47618. doi: 10.18632/oncotarget.17744
Ryall, J. G., Cliff, T., Dalton, S., and Sartorelli, V. (2015). Metabolic reprogramming of stem cell epigenetics. Cell Stem Cell 17, 651–662. doi: 10.1016/j.stem.2015.11.012
Sato, N., Meijer, L., Skaltsounis, L., Greengard, P., and Brivanlou, A. H. (2004). Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 10, 55–63. doi: 10.1038/nm979
Shiraki, N., Shiraki, Y., Tsuyama, T., Obata, F., Miura, M., Nagae, G., et al. (2014). Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 19, 780–794. doi: 10.1016/j.cmet.2014.03.017
Shyh-Chang, N., Locasale, J. W., Lyssiotis, C. A., Zheng, Y., Teo, R. Y., Ratanasirintrawoot, S., et al. (2013). Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 339, 222–226. doi: 10.1126/science.1226603
Sperber, H., Mathieu, J., Wang, Y., Ferreccio, A., Hesson, J., Xu, Z., et al. (2015). The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat. Cell Biol. 17, 1523–1535. doi: 10.1038/ncb3264
Szulc, J., Wiznerowicz, M., Sauvain, M. O., Trono, D., and Aebischer, P. (2006). A versatile tool for conditional gene expression and knockdown. Nat. Methods 3, 109–116. doi: 10.1038/nmeth846
Teslaa, T., and Teitell, M. A. (2015). Pluripotent stem cell energy metabolism: an update. EMBO J. 34, 138–153. doi: 10.15252/embj.201490446
Thomas, G., Betters, J. L., Lord, C. C., Brown, A. L., Marshall, S., Ferguson, D., et al. (2013). The serine hydrolase ABHD6 is a critical regulator of the metabolic syndrome. Cell Rep. 5, 508–520. doi: 10.1016/j.celrep.2013.08.047
Tzelepis, K., Koike-Yusa, H., De Braekeleer, E., Li, Y., Metzakopian, E., Dovey, O. M., et al. (2016). A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 17, 1193–1205. doi: 10.1016/j.celrep.2016.09.079
Vijayakumar, A., Vijayaraj, P., Vijayakumar, A. K., and Rajasekharan, R. (2016). The Arabidopsis ABHD11 mutant accumulates polar lipids in leaves as a consequence of absent acylhydrolase activity. Plant Physiol. 170, 180–193. doi: 10.1104/pp.15.01615
Wang, J., Alexander, P., Wu, L., Hammer, R., Cleaver, O., and McKnight, S. L. (2009). Dependence of mouse embryonic stem cells on threonine catabolism. Science 325, 435–439. doi: 10.1126/science.1173288
Wang, L., Zhang, T., Wang, L., Cai, Y., Zhong, X., He, X., et al. (2017). Fatty acid synthesis is critical for stem cell pluripotency via promoting mitochondrial fission. EMBO J. 36, 1330–1347. doi: 10.15252/embj.201695417
Wiedl, T., Arni, S., Roschitzki, B., Grossmann, J., Collaud, S., Soltermann, A., et al. (2011). Activity-based proteomics: identification of ABHD11 and ESD activities as potential biomarkers for human lung adenocarcinoma. J. Proteomics 74, 1884–1894. doi: 10.1016/j.jprot.2011.04.030
Wu, J., and Belmonte, J. C. I. (2016). Stem cells: a renaissance in human biology research. Cell 165, 1572–1585. doi: 10.1016/j.cell.2016.05.043
Yanes, O., Clark, J., Wong, D. M., Patti, G. J., Sanchez-Ruiz, A., Benton, H. P., et al. (2010). Metabolic oxidation regulates embryonic stem cell differentiation. Nat. Chem. Biol. 6, 411–417. doi: 10.1038/nchembio.364
Yang, J., Takahashi, Y., Cheng, E., Liu, J., Terranova, P. F., Zhao, B., et al. (2010). GSK-3beta promotes cell survival by modulating Bif-1-dependent autophagy and cell death. J. Cell Sci. 123(Pt 6), 861–870. doi: 10.1242/jcs.060475
Ying, Q. L., Wray, J., Nichols, J., Batlle-Morera, L., Doble, B., Woodgett, J., et al. (2008). The ground state of embryonic stem cell self-renewal. Nature 453, 519–523. doi: 10.1038/nature06968
Young, M. M., Kester, M., and Wang, H. G. (2013). Sphingolipids: regulators of crosstalk between apoptosis and autophagy. J. Lipid Res. 54, 5–19. doi: 10.1194/jlr.R031278
Zechner, R., Zimmermann, R., Eichmann, T. O., Kohlwein, S. D., Haemmerle, G., Lass, A., et al. (2012). FAT SIGNALS–lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 15, 279–291. doi: 10.1016/j.cmet.2011.12.018
Zhang, H., Badur, M. G., Divakaruni, A. S., Parker, S. J., Jager, C., Hiller, K., et al. (2016). Distinct metabolic states can support self-renewal and lipogenesis in human pluripotent stem cells under different culture conditions. Cell Rep. 16, 1536–1547. doi: 10.1016/j.celrep.2016.06.102
Zhang, J., Liu, G., Ruan, Y., Wang, J., Zhao, K., Wan, Y., et al. (2014). Dax1 and Nanog act in parallel to stabilize mouse embryonic stem cells and induced pluripotency. Nat. Commun. 5:5042. doi: 10.1038/ncomms6042
Zhang, J., Nuebel, E., Daley, G. Q., Koehler, C. M., and Teitell, M. A. (2012). Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell 11, 589–595. doi: 10.1016/j.stem.2012.10.005
Zhang, J., Ratanasirintrawoot, S., Chandrasekaran, S., Wu, Z., Ficarro, S. B., Yu, C., et al. (2016). LIN28 regulates stem cell metabolism and conversion to primed pluripotency. Cell Stem Cell 19, 66–80. doi: 10.1016/j.stem.2016.05.009
Keywords: ABHD11, embryonic stem cell, self-renewal, overexpression, knockout
Citation: Liu G, Ruan Y, Zhang J, Wang X, Wu W, He P, Wang J, Xiong J, Cheng Y, Liu L, Yang Y, Tian Y and Jian R (2020) ABHD11 Is Critical for Embryonic Stem Cell Expansion, Differentiation and Lipid Metabolic Homeostasis. Front. Cell Dev. Biol. 8:570. doi: 10.3389/fcell.2020.00570
Received: 11 March 2020; Accepted: 15 June 2020;
Published: 07 July 2020.
Edited by:
Karthikeyan Narayanan, West Virginia University, United StatesReviewed by:
Shannon Buckley, University of Nebraska Medical Center, United StatesTetsuya S. Tanaka, Elixirgen Scientific, Inc., United States
Copyright © 2020 Liu, Ruan, Zhang, Wang, Wu, He, Wang, Xiong, Cheng, Liu, Yang, Tian and Jian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanping Tian, tianyp1981@163.com; Rui Jian, jianruilq2@aliyun.com
†These authors have contributed equally to this work
 Gaoke Liu
Gaoke Liu Yan Ruan1†
Yan Ruan1†  Wei Wu
Wei Wu