Abstract
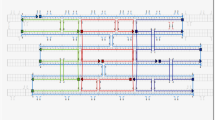
Since less than a decade ago, the DNA origami technique has become an important tool in nanopore fabrication. DNA origami nanopores are highly efficient because of their compatibility with biomolecules and the possibility to precisely engineer their dimensions and designs. However, accurate comprehension of their molecular behavior under various conditions is still unsatisfactory. In this study, a thin plate DNA origami nanopore is designed and investigated using molecular dynamics simulation. The thin plate is designed using caDNAno software along with the square lattice method and the molecular dynamics simulation is performed using GROMACS software. The model is simulated in a wide temperature range and its stability is investigated. The shape and dimensions of the nanopore are also compared at these temperatures. The results indicate that the designed nanopore exhibits decent stability at these temperatures and no breakdown was observed despite some distortions in the structure at high temperatures. In addition, the effect of the number of staple strands on the structure, stability, and deformation of the DNA origami plate is investigated and it is found that addition of staple strands have a significant positive effect on the stability of nanopore’s shape. By the results of analyzing the shape of the nanopore, it suggests that the proposed nanopore can be used to pass a wide range of molecules, macromolecules, and drug cargoes.










Similar content being viewed by others
References
Seeman, N. C. (1982). Nucleic acid junctions and lattices. Journal of Theoretical Biology,99, 237–247.
Watson, J. D., & Crick, F. H. C. (1953). The structure of DNA. Cold Spring Harbor Laboratory Press,18, 123–131.
Rothemund, P. W. K. (2006). Folding DNA to create nanoscale shapes and patterns. Nature,440, 297–302.
Douglas, S. M., Dietz, H., Liedl, T., Högberg, B., Graf, F., & Shih, W. M. (2009). Self-assembly of DNA into nanoscale three-dimensional shapes. Nature,459, 414–418.
Dietz, H., Douglas, S. M., & Shih, W. M. (2009). Folding DNA into twisted and curved nanoscale shapes. Science,325, 725–730.
Han, D., Pal, S., Nangreave, J., Deng, Z., Liu, Y., & Yan, H. (2011). DNA origami with complex curvatures in three-dimensional space. Science,332, 342–346.
Pinheiro, A. V., Han, D., Shih, W. M., & Yan, H. (2011). Challenges and opportunities for structural DNA nanotechnology. Nature Nanotechnology,6, 763–772.
Tørring, T., Voigt, N. V., Nangreave, J., Yan, H., & Gothelf, K. V. (2011). DNA origami: A quantum leap for self-assembly of complex structures. Chemical Society Reviews,40, 5636–5646.
Castro, C. E., Kilchherr, F., Kim, D.-N., Shiao, E. L., Wauer, T., Wortmann, P., et al. (2011). A primer to scaffolded DNA origami. Nature Methods,8, 221–229.
Kuzuya, A., & Komiyama, M. (2010). DNA origami: Fold, stick, and beyond. Nanoscale,2, 310–322.
Kasianowicz, J. J., Brandin, E., Branton, D., & Deamer, D. W. (1996). Characterization of individual polynucleotide molecules using a membrane channel. Proceedings of the National Academy of Sciences of United States of America,93, 13770–13773.
Branton, D., Deamer, D. W., Marziali, A., Bayley, H., Benner, S. A., Butler, T., et al. (2008). The potential and challenges of nanopore sequencing. Nature Biotechnology,26, 1146–1153.
Pennisi, E. (2012). Search for pore-fection. Science,336, 536–537.
Kasianowicz, J. J., Brandin, E., Branton, D., & Deamer, D. W. (1996). Characterization of individual polynucleotide molecules using a membrane channel. Proceedings of the National academy of Sciences of the United States of America,93, 13770–13773.
Bayley, H., & Martin, C. (2000). Resistive-pulse sensing-from microbes to molecules. Chemical Reviews,100, 2575–2594.
Deamer, D. W., & Branton, D. (2002). Characterization of nucleic acids by nanopore analysis. Accounts of Chemical Research,35, 817–825.
Song, L., Hobaugh, M. R., Shustak, C., Cheley, S., Bayley, H., & Gouaux, J. E. (1996). Structure of staphylococcal a-hemolysin, a heptameric transmembrane pore. Science,274, 1859–1866.
Garaj, S., Hubbard, W., Reina, A., Kong, J., Branton, D., & Golovchenko, J. A. (2010). Graphene as a subnanometre trans-electrode membrane. Nature,467, 190–193.
Wei, R., Martin, T. G., Rant, U., & Dietz, H. (2012). DNA origami gatekeepers for solid-state nanopores. Angewandte Chemie International Edition,51, 4864–4867.
Bell, N. A. W., Engst, C. R., Ablay, M., Divitini, G., Ducati, C., Liedl, T., et al. (2012). DNA origami nanopores. Nano Letters,12, 512–517.
Krishnan, S., Ziegler, D., Arnaut, V., Martin, T. G., Kapsner, K., Henneberg, K., et al. (2016). Molecular transport through large-diameter DNA nanopores. Nature Communications.,7, 12787.
Plesa, C., Ananth, A. N., Linko, V., et al. (2014). Ionic permeability and mechanical properties of DNA origami nanoplates on solid-state nanopores. American Chemical Society,8, 35–43.
Kim, D. N., Kilchherr, F., Dietz, H., & Bathe, M. (2012). Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Research,40, 2862–2868.
Ouldridge, T. E., Louis, A. A., & Doye, J. P. K. (2010). DNA nanotweezers studied with a coarse-grained model of DNA. Physical Review Letters,104, 178101.
Ouldridge, T. E., Louis, A. A., & Doye, J. P. K. (2011). Structural, mechanical, and thermodynamic properties of a coarse-grained DNA model. The Journal of Chemical Physics,134, 085101.
Snodin, B. E. K., Randisi, F., Mosayebi, M., Sulc, P., Schreck, J. S., Romano, F., et al. (2015). Introducing improved structural properties and salt dependence into a coarse-grained model of DNA. The Journal of Chemical Physics,142, 234901.
Yoo, J., & Aksimentiev, A. (2013). In situ structure and dynamics of DNA origami determined through molecular dynamics simulations. Proceedings of the National Academy of Sciences of the United States of America,110, 20099–20104.
Maffeo, C., Yoo, J., & Aksimentiev, A. (2016). De novo reconstruction of DNA origami structures through atomistic molecular dynamics simulation. Nucleic Acids Research,44(7), 3013–3019.
Li, C. Y., Hemmig, E. A., Kong, J., Yoo, J., Hernandez-Ainsa, S., Keyser, U. F., et al. (2015). Ionic conductivity, structural deformation, and programmable anisotropy of dna origami in electric field. ACS Nano,9(2), 1420–1433.
Yoo, J., & Aksimentiev, A. (2015). Molecular dynamics of membrane-spanning DNA channels: Conductance mechanism, electro-osmotic transport, and mechanical gating. Journal of Physical Chemistry Letters,6, 4680–4687.
Göpfrich, K., Li, C. Y., Mames, I., Bhamidimarri, S. P., Ricci, M., Yoo, J., et al. (2016). Ion channels made from a single membrane-spanning DNA duplex. Nano Letters,16, 4665–4669.
Göpfrich, K., Li, C. Y., Ricci, M., Bhamidimarri, S. P., Yoo, J., Gyenes, B., et al. (2016). Large-conductance transmembrane porin made from DNA origami. ACS Nano,10, 8207.
Ohmann, A., Li, C. Y., Maffeo, C., Al Nahas, K., Baumann, K. N., Göpfrich, K., et al. (2018). A synthetic enzyme built from DNA flips 107 lipids per second in biological membranes. Nature Communication,9, 2426.
Belkin, M., & Aksimentiev, A. (2016). Molecular dynamics simulation of DNA capture and transport in heated nanopores. ACS Applied Materials,8, 12599–12608.
Douglas, S. M., Marblestonen, A. H., Teerapittayanon, S., Vazquez, A., Church, G. M., & Shih, W. M. (2009). Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Research,37(15), 5001–5006.
Ramakrishnan, S., Ijäs, H., Linko, V., & Keller, A. (2018). Structural stability of DNA origami nanostructures under application specific conditions. Computational and Structural Biotechnological Journal,16, 342–349.
Pillers, M. A., & Lieberman, M. (2014). Thermal stability of DNA origami on mica. Journal of Vacuum Science and Technology,32, 040602.
Rajendran, A., Endo, M., Katsuda, Y., Hidaka, K., & Sugiyama, H. (2011). Photo-cross linking assisted thermal stability of DNA origami structures and its application for higher-temperature self-assembly. Journal of the American Chemical Society,133(37), 14488–14491.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Khosravi, R., Ghasemi, R.H. & Soheilifard, R. Design and Simulation of a DNA Origami Nanopore for Large Cargoes. Mol Biotechnol 62, 423–432 (2020). https://doi.org/10.1007/s12033-020-00261-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-020-00261-z