Abstract
Amplicon-based next generation sequencing (NGS) approaches have been preferentially adopted by the clinical laboratories on the basis of a short turnaround time (TAT) and small DNA input needs. However, little work has been done to assess the amplicon-based NGS methods for copy number variation (CNV) detection in comparison with current standard methods like immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH). The correlation between NGS based CNV detection and the later standard methods has remained unexplored. We developed an amplicon-based panel to detect human epidermal receptor growth factor (HER2) amplification in formalin-fixed paraffin-embedded (FFPE) tumor tissue samples from 280 breast cancer and 50 gastric cancer patients. Assessment by IHC and FISH was conducted in parallel, and descriptive statistics were used to assess the concordance. The copy number detected by NGS was correlated with either the average HER2 copy number (signals/cell) (r = 0.844; p < 0.001) or the HER2/CEP17 ratio (r = 0.815; p < 0.001). We determined a cut-off value for NGS to categorize HER2 amplification status by using 151 HER2 non-amplified FFPE samples. In breast cancer patients, the cut-off value was 2.910, with 95.35%, 98.67% and 97.29% sensitivity, specificity and concordance, respectively. However, this cut-off value displayed low sensitivity in gastric cancer patients (64.71%), and the following macrodissection procedure was not effective for increasing sensitivity (57.14%). Evaluation of HER2 copy number with NGS in our study was comparable with IHC and FISH in breast cancer patients, but concordance in gastric cancer was only moderate. The greater discordance in gastric cancer may reflect the underlying biological mechanisms, and further study is warranted. NGS-based HER2 assessment may decrease the equivocal HER2 determinations in breast cancer patients assessed by FISH/IHC.
Similar content being viewed by others
Background
The reliable identification of clinically actionable genomic alteration method is critical for the precision cancer therapy guidance. Next generation sequencing (NGS) has the capability to simultaneously assess multiple genes with a limited biopsy material, thus representing both a cost and tissue-efficient alternative to current single-gene assessment methods [1,2,3]. Prior studies have shown that NGS enables a reliable detection for copy number variations (CNV) from the same assays used to detect sequence alterations, but less information is available on amplicon-based target sequences [4,5,6,7]. CNV calling in the amplicon sequence relies on the calculation of amplicon coverage and suitable normalization. Several factors influence CNV detection, including the number of amplicons per gene, average read dept., and tumor purity within the sample [7]. Thus, an assay and algorithm shall need to be fully validated before being clinically used.
Human epidermal receptor growth factor (HER2) amplification or an overexpression occurs in approximately 18–20% of breast cancers, and in nearly 20% of gastric or gastroesophageal junction (GEJ) cancers [8, 9]. Several methods have been recommended for HER2 amplification assessment, including in situ hybridization (ISH) techniques, which evaluate HER2 status by measuring the number of HER2 gene copies, or IHC, which quantifies protein expression [9]. The ASCO/CAP has provided detailed guidelines for conducting and interpreting HER2 status in a clinical practice. These scoring methods classify cases into “positive”, “negative”, and “equivocal” categories [10]. According to these guidelines, equivocal HER2 status necessitates additional testing, thus increasing the cost of patient management, and delaying the decision to recommend HER2-targeted therapy. The correlation between a copy number called by NGS with an average HER2 copy number, HER2/CEP17 ratio, or IHC score is not well established. With increasing NGS use in a clinical practice it is increasingly important to validate the amplicon-based detection method against the standard methodologies.
In the current study, we performed and evaluated an amplicon-based NGS assay to assess HER2 amplification in breast and gastric cancers, by using a custom designed panel and bioinformatics pipeline. We evaluated accuracy and concordance of NGS detection compared with the gold-standard FISH/IHC analysis methodologies.
Materials and Methods
Study and Panel Design
For CNV detection, we designed an amplicon-based panel covering 50 genes, which included 13 CNV genes and 6 baseline genes (supplement Tables 1, 2). Briefly, we used cell lines with known amplifications for validating accuracy and had great precision for copy number detection. Then, it was expanded to the FFPE samples from both the breast and gastric cancer patients. We further compared the copy number detected by NGS with the FISH/IHC results from the same sample, and determined a cut-off value of NGS to determine HER2 status. The study schema is summarized in the supplementary Fig. 1.
Cell Line CNV Detection
To validate CNV detection, we pooled four cell lines, each bearing single focal gene amplification (HER2, MET, EGFR and FGFR3) with the matched normal cell lines (GM18511) in several dilution series (40%, 30%, 25%, 20%, 15%, 10%, 5%, 4.5%, 3%). The standard materials list is shown in supplementary Table 3. For orthogonal support, the copy number of the molecular standard materials were also measured by digital PCR using the QuantStudio 3D digital PCR system (Life Technology, CA, USA).
Patients and Samples
To verify our custom designed 50 gene panel in clinical application, we used FFPE samples from 280 invasive breast cancer patients and 50 gastric cancer patients obtained from Peking University Cancer Hospital. Five FFPE slides, each 5-μm thick, were obtained from breast cancer patient samples along with ten FFPE slides from gastric cancer. Tumors with a high degree of necrosis and < 1000 tumor cells were excluded. More than 80% of the samples we finally selected were samples with tumor purity greater than 20%. This study was approved by the Medical Ethics Committee of Peking University Cancer Hospital, and the investigation was performed in accordance with the Declaration of Helsinki Principles. All patients had signed informed consent for the tissue research, and all of clinical data and samples were deidentified prior to analysis. All the experiments were carried out in accordance with the guideline released by the National Health and Family Planning Commission of the PRC.
DNA Extraction
Genomic DNA was extracted from unstained FFPE samples using TIANamp FFPE DNA Kit (TIANGEN, Beijing, China), according to manufacturer’s instructions. DNA was quantified using the Qubit dsDNA HS Assay Kit (Life Technology, CA, USA) and the Qubit 2.0 Flurometer (Life Technology, CA, USA) according to recommended protocols. Quality checks were performed by testing 5 ng DNA in 1% agarose gel electrophoresis. Samples in which the main DNA strip in agarose gel electrophoresis less than 600 bp were excluded. The DNA was stored at −20 °C.
NGS Library Preparation
Sequence libraries were prepared by using library preparation reagents from Life Technology, CA, USA. The amount of DNA input was 15 ng. Libraries were constructed using a custom designed panel (50 hotspot genes). Then, the amplicons by Ion Ampliseq Library Kit 2.0 were barcoded during library generation using the Ion Xpress Barcode Adapters 1–96 Kit. The libraries were purified by AMPure XP beads, quantified using the Ion Library Quantitation Kit, and qualified using Agilent Bioanalyzer 2100. Then the libraries were pooled for sequencing. Multiplex barcoded libraries were enriched by clonal amplification using emulsion PCR on Ion Sphere particles (Ion PI™ Template OT2 200 Kit v3, Life Technology, CA, USA) and loaded on an Ion PI™ Chip. Massively parallel sequencing was carried out on Ion Proton platform using the Ion PI™ Sequencing 200 Kit v3 according to manufacturer’s instructions.
Sequencing and Data Analysis
Torrent Suite Software (version 4.4.3) was used to perform signal processing, base calling, quality score assignment, and adapter trimming after the sequencing reaction. High quality reads were aligned to human genome 19 reference by tmap4.2.18 software. Quality control and coverage analysis was performed by an in-house analysis pipeline.
Base Substitution, Short Insertion and Deletion Analysis
TVC (Torrent Variant Caller, version 4.4) was used to call SNV and InDel variants. TVC modules use freebayes to discover candidate variants combined with the hotspots file for detecting gene mutations. Somatic mutations were determined using the following filters: (i) the minimum coverage depth was 100 for SNP and 200 for InDel; (ii) the minimum cutoff of MAF was 0.01 for hotspot variants and 0.05 for others; (iii) detected SNVs and InDels also required at least 25 variant-contained reads to be reported as positive. Those combined minimum coverage, MAF and variant-contained reads to ensure the accuracy of variant calls.
Copy Number Variation Detection
We used an exome-like approach, rather than the average coverage of exon pull-down regions with read counts per amplicon, for identifying CNV. The coverage of each amplicon was calculated as the number of reads which covered more than one amplicon but mostly aligned to the amplicon. Then, the amplicon-level coverage was divided by the median coverage of each amplicon to normalize or minimize inter-sample variation. The normalized amplicon-level coverage was also corrected by GC content to remove the dependency of coverage across the different GC profiles. Amplicons with a coverage of less than 100 × were excluded from analysis. The copy number ratio of each amplicon was calculated by dividing the GC corrected amplicon-level coverage of tumor samples with that of the matched normal sample or normal pool. In this study, we used a normal pool derived from 14 normal breast cancer patient FFPE samples instead of the matched normal sample for a reference of diploid genome comparison. The copy number ratio was then log-transformed to yield the log2 copy number ratio, which was subsequently used to determine gene amplification status. The gene level fold change was determined as the weighted average of amplicon-level log-copy number ratios, for which the weight of each amplicon was proportional to the number of reads; basically, the reads in the matched normal samples or the normal pool. The final copy number of gene was equal to twice the gene level fold change.
HER2 IHC and FISH Testing
HER2 amplification was determined by IHC and the dual probe FISH test. FISH results were reported as average HER2 copy number and HER2/CEP17 ratio. All FFPE samples were reviewed by two individual pathologists to determine HER2 status.
Statistics
The accuracy (sensitivity and specificity) and precision (repeatability and reproducibility) of NGS was evaluated with standard material result. The correlations of the copy number called by NGS and that determined by digital PCR were studied by using R software. Comparisons of copy numbers which were detected in the three runs, were analyzed using the ANOVA test.
Results
Assay Performance of NGS Calls in Standard Materials
The NGS assay performance for detecting CNV was analyzed by detecting standard materials. The copy number of standard materials is summarized in Supplementary Table 3. The precision was assessed in inter-assay and intra-assay studies. We first simultaneously ran the two libraries which had been prepared by two different operators (Lib1 and Lib3). Then, Lib1 was done on another run (Lib2), yielding a total of 3 replicates for each sample. Then, it was repeatability evaluated on a per-gene basis among the replicates. Each gene had also a similar copy number estimation in the replicated libraries (Fig. 1). No statistical differences in the copy number could be detected among the three runs (F-value = 0.022, P value = 0.9783). The coefficient of variation (CV%) for the variation in copy number was <8.58% for inter-assay, and 8.77% for inner-assay, respectively. The total CV% for the three replicates ranged from 0.49% to 7.33% (Supplementary Table 4). A high specimen’s correlation was detected when we compared the copy number for all evaluable genes on the targeted panel with the one generated with digital PCR (Fig. 2a). We also assessed the effect of tumor purity on sensitivity for CNV detection by preparing libraries from tumor DNA diluted with varying amounts of normal DNA, observing that the copy number was linearly related to tumor purity. The level of amplification also greatly impacted the CNV detection using NGS. Genes with a high CNV could be detected with a lower tumor purity than those with a low CNV (Fig. 2b). Subsequent analyses were focused specifically on HER2 detection.
The correlation analysis of CNV detection by preparing libraries. a Curve regression analysis between copy number detected by NGS and digital PCR. The copy number detected by digital PCR was used as expected copy number. The copy number detected by NGS was used as detected copy number. b The effect of tumor purity on the detection limit for gene amplifications. Four cell lines each bearing single focal gene amplification (HER2, MET, EGFR and FGFR3) were pooled with their matched normal cell lines in several dilution series. Detection limits were evaluated with dilution series
Distribution of HER2 Amplification Status in Breast Cancer
All the 280 FFPE samples from breast cancer patients were tested using FISH and IHC for establishing the HER2 amplification status. The IHC results showed that 31.4% (88/280) scored as IHC 3+, 185 as IHC 2+ and 7 as IHC 1+. The specimens were then tested by FISH, except for one IHC 2+ sample and one IHC 3+ sample, for which testing had failed. FISH results were 100% consistent with IHC for 1+ and 3+ samples. For 184 IHC 2+ samples, a total of 121 negative samples, 63 positive samples. Two pathologists reviewed these histologic findings, and ranked 143 samples as negative and 41 samples as positive. Overall, this led to 129 positive samples, 151 negative samples.
Determination and Evaluation of Cut-off to Categorize the HER2 Status in Breast Cancer Using NGS
After the validation in the cell lines, we implemented CNV detection in large scale clinical FFPE specimens. The NGS-detected copy number was correlated with the FISH results (Fig. 3). To assess the sensitivity for detecting CNV when compared to the gold-standard method, we evaluated the quantitative correlation between NGS and the average HER2 copy number or HER2/CEP17 ratio detected by FISH. We used a total of 255 samples to fit a linear regression model of NGS copy number and the FISH testing results (Supplementary Table 5), excluding those with FISH testing failure (n = 3), HER2 status reviewed by pathologists (n = 22). The log10 copy number detected by NGS correlated with either the log10 average HER2 copy number (signals/cell) (y = 0.044 + 0.73x, r = 0.844; add p < 0.001), and log10 HER2/CEP17 ratio (y = 0.26 + 0.74x, r = 0.815; add p < 0.001). As NCCN guideline considered average copy number of HER2 ≥ 6.0 signals/cell as positive, if the average copy number is 6 then the NGS copy number is 4.09 according to this equation. Similarly, if the average copy number is 4, the NGS copy number will be 3.04, and if the HER2/CEP17 ratio is set as 2, the NGS copy number will be 3.03. No false-positive or false-negative samples were found when the copy number detected by NGS was >4.09. This value was hence identified as the confident positive cut-off.
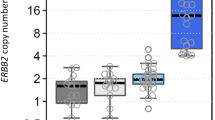
Amplicon-based NGS to detect CNV from breast cancer specimens identified by IHC and FISH. Representative microscopical results of HER2 amplification (positive, negative and equivocal HER2 status) were shown by IHC (a, d, g) and dual color FISH (b, e, h). FFPE resection specimens with identified by FISH were further analyzed by NGS (c, f, i). The y-axis shows log2 copy number ratios of each amplicon from each gene
To determine the negative cut-off value, we used 151 HER2 negative samples to represent the copy number distribution in HER2 negative samples. The copy number data followed a normal distribution (P value of Shapiro-Wilk test = 0.0539). We then used the mean + 3 × MAD (mean absolute deviation) as the negative cut-off value (Supplementary Fig. 2). Two false-positive sample was found when the copy number ranged between 2.91 and 4.09, and five false-negative samples were observed when the copy number was <2.91. Excellent performance was hence found for amplification status when NGS copy number was >4.09 and < 2.91, and the copy number between 2.91 and 4.09 as weak positive status. Overall, we achieved a sensitivity of 95.35% (123/129) and a specificity of 98.67% (149/151) compared with HER2 status, which was determined by two pathologists who considered both IHC and FISH testing results in breast cancer (Table 1).
Classification of HER2 Equivocal Status Samples in Breast Cancer Using NGS
Compared with the HER2 status by IHC score, all 7 IHC 1+ samples were classified as HER2-negative by NGS. Among 88 IHC 3+ samples, one (1/88) was identified to be negative by NGS (copy number = 2.71). For 185 IHC 2+ samples, NGS had an 87.80% (36/41) sensitivity and a 98.61% (142/144) specificity compared with HER2 status determined by two pathologists after considering both IHC and FISH testing.
Seven discrepancies were found, six of which had occurred in the context of a lowered tumor content and HER2 heterogeneity (Supplementary Fig. 3). The average HER2 copy number of these samples was 10.6, 5.9, 5.1, 4.1, 13.48 and 7.5, respectively. The HER2/CEP17 ratio was 3.2, 2.9, 4.7, 2.28, 6.1 and 4.4, respectively. However, the copy number of these samples were < 2.91. All the six specimens displayed high tumor heterogeneity, which was thought to influence NGS accuracy.
CNV Detection in Gastric Cancer Using NGS
A total number of 50 gastric cancer patients were enrolled in this study. In the initial 39 fully extracted FFPE samples, NGS identified 11 out of 17 HER2 positive samples, and all the 22 HER2 negative samples. Although the positive predictive value (PPV) was 100%, NGS categorized all HER2 negative samples correctly, with a low sensitivity (64.71%, 11/17). Compared with breast cancer, gastric cancer appeared to be more heterogeneous. We hence decided to evaluate whether a macrodissection based IHC result would help to increase the sensitivity. There were 28 HER2 amplified samples sequenced after macrodissection, and sensitivity was 57.14% (16/28). Although the NGS-detected copy number of most samples increased after macrodissection (Fig. 4), it was not sufficient to improve the accuracy of the NGS assay for HER2 detection in gastric cancer (Table 2).
Effect of macrodissection on HER2 detection by NGS in gastric cancer. a Diagram showing macrodissection of FFPE tissue area of HE stained section of gastric cancer. b Serial section from the same specimen for HER2 IHC. c HER2 FISH (red signal, HER2; green signal, CEP17). d CNV detection by NGS after macrodissection. e CNV detection by NGS before macrodissection
Concurrent Detection of Other Somatic Alterations in the Clinical FFPE Samples
Overall, 118 breast cancer samples with at least one gene mutation detected were include in this study. The most common mutations were PIK3CA (109/280), TP53 (4/280), AKT1 (3/280), KRAS (3/280), HER2 (2/280), ALK (1/280), EGFR (1/280), and RET (1/280). Among these mutations, annotated based on the OncoKb Knowledge Base [11], PIK3CA, AKT1 and HER2 mutations were labelled with a 3A level (Compelling clinical evidence supports the biomarker as being predictive of response to a drug in this indication, but neither biomarker nor drug are standard care). When assigning samples to the level of the most actionable alteration, 40.71% (114/280) patients harbored at least one potentially actionable alteration, which may be a response to a drug, although this is not been defined as standard care so far.
In gastric cancer, MSH3 was the most common mutation, occurring in 74% (37/50) patients. The frequency of this mutation was higher in gastric cancer without HER2 amplifications (HER2-negative patients: 91%, HER2-positive patients: 61%; p = 0.036, Chi-square test).
Discussion
HER2 amplification has both predictive and prognostic value for breast cancer. Currently, it is regarded as the only biomarker established for selecting specific therapy for patients with advanced gastric cancer [12, 13]. The current gold-standard approach for assessing HER2 amplification status is based on the IHC and ISH techniques. However, debate continues over the best way to relate HER2 test results with treatment outcomes. One drawback of the HER2 test is that the scoring system used to determine HER2 status is subjective. In this study, we developed an amplicon-based NGS panel to accurately detect clinically relevant copy number alterations. Evaluating HER2 copy number with NGS in our study yielded comparable results to the gold-standard FISH/IHC analyses in breast cancer patients, achieving 95.35% sensitivity and 98.67% specificity.
The cell line dilution study showed that the detection for amplification is strongly influenced by tumor purity. As shown in Fig. 2b, copy number was reduced in parallel with decrease of tumor purity. For example, the initial copy number was about 4 for FGFR1 at 80% tumor purity, whilst the copy number was approximately 2.45 at 20% tumor purity. Thus, if the initial amplification level was low, it would not be detectable at a low tumor purity. Other studies also shown that samples with poor quality or low DNA content can yield noisy CN plots, thus limiting accurate assessment. The performance was also affected with lower CNVs (6–7 copies) and in samples with poor purity (20–30%) [14]. Thus, determining adequate tumor purity would be mandatory, for an accurate assessment. Furthermore, an alternate method shall be recommended for cases with amplification in combination with low tumor content.
In addition to providing an accurate copy number, we also need to transfer the continuous copy number value into a binary amplification status. In this study, we used known negative samples to determine the negative cut-off value of NGS and established that 2.91 yielded a 95.35% sensitivity and 98.67% specificity when compared to the gold-standard method. To be more confident with NGS CNV detection, we used the established cut-off value for FISH, to estimate a positive cut-off value of NGS according to their correlation. It showed that a copy number using NGS higher than 4.09 corresponded to an average HER2 copy number higher than 6 signs/cell. Thus, we recommended an NGS copy number between 2.91 and 4.09 to take an additional reflex test. However, it could be foreseen that larger validated samples would reduce this greyscale. In fact, our methods derive a nearly identical cutoff to call HER2 amplificated by NGS as the large commercial NGS provided Foundation Medicine, Inc. which uses 4 NGS-derived copies to call amplification in HER2.
According to updated 2018 ASCO/CAP guideline, concomitant IHC assays are required to arrive at the most accurate HER2 status designation after HER2 FISH equivocal results. Currently, our research gave preliminary suggestions, whether dual-probe ISH group 2 to 4 in 2018 ASCO/CAP guideline can be considered for inclusion in the negative. NGS might provide accurate assessment for the HER2 status designation, and thus reduces the risk of misdiagnosis, and further verification is required.
NGS still has some shortcomings for detection mutations, e.g., CNV detection accuracy was based on accurately assessing coverage depth of genes, which can be biased by high GC content and repetitive regions. A previous study reported that the number of amplicons per gene on the panel may influence performance of CNV detection [7]. The higher number of amplicons per gene would have somehow decreased the variance in CNV assessment, but it would have also restricted the list of assessable genes.
In some cases, metastatic tumors have different molecular alterations from the primary tumors. Even in breast cancer, a discrepancy of HER2 status between primary tumor and distant metastases has been observed in 7–26% of patients [15]. Regarding gastric cancer, tumor heterogeneity could be precisely identified using ctDNA [16] or planning reflex testing on residual materials or additional tumor blocks. Similar findings have been reported in advanced gastric cancer patients, whose primary tumors were found to be HER2 negative, but whose circulating tumor cells displayed HER2 amplification [17].
Conclusions
Our study demonstrate that an optimized NGS-based test can accurately detect most clinically targetable CNV in a broad spectrum of cancer patients. NGS-based HER2 assessment may decrease the equivocal HER2 determinations in breast cancer patients assessed by FISH/IHC. However, due to heterogeneity of gastric cancer tumor tissue, detection of HER2 amplification by NGS seems still problematic in this malignancy.
Abbreviations
- NGS:
-
next generation sequencing
- CNV:
-
copy number variation
- IHC:
-
immunohistochemistry
- FISH:
-
fluorescence in situ hybridization
- HER2 :
-
human epidermal receptor growth factor
- FFPE:
-
formalin-fixed paraffin-embedded
References
Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, Paz-Ares L (2017) Lung cancer: current therapies and new targeted treatments. Lancet 389(10066):299–311. https://doi.org/10.1016/S0140-6736(16)30958-8
Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, Deignan JL, Friez MJ, Funke BH, Hegde MR, Lyon E, Working Group of the American College of Medical G, Genomics Laboratory Quality Assurance C (2013) ACMG clinical laboratory standards for next-generation sequencing. Genet Med 15(9):733–747. https://doi.org/10.1038/gim.2013.92
Hamblin A, Wordsworth S, Fermont JM, Page S, Kaur K, Camps C, Kaisaki P, Gupta A, Talbot D, Middleton M, Henderson S, Cutts A, Vavoulis DV, Housby N, Tomlinson I, Taylor JC, Schuh A (2017) Clinical applicability and cost of a 46-gene panel for genomic analysis of solid tumours: retrospective validation and prospective audit in the UK National Health Service. PLoS Med 14(2):e1002230. https://doi.org/10.1371/journal.pmed.1002230
Pfarr N, Penzel R, Endris V, Lier C, Flechtenmacher C, Volckmar AL, Kirchner M, Budczies J, Leichsenring J, Herpel E, Noske A, Weichert W, Schneeweiss A, Schirmacher P, Sinn HP, Stenzinger A (2017) Targeted next-generation sequencing enables reliable detection of HER2 (ERBB2) status in breast cancer and provides ancillary information of clinical relevance. Gene Chromosome Cancer 56(4):255–265. https://doi.org/10.1002/gcc.22431
Yamamoto T, Shimojima K, Ondo Y, Imai K, Chong PF, Kira R, Amemiya M, Saito A, Okamoto N (2016) Challenges in detecting genomic copy number aberrations using next-generation sequencing data and the eXome hidden Markov model: a clinical exome-first diagnostic approach. Hum Genome Var 3:16025. https://doi.org/10.1038/hgv.2016.25
Ellingford JM, Campbell C, Barton S, Bhaskar S, Gupta S, Taylor RL, Sergouniotis PI, Horn B, Lamb JA, Michaelides M, Webster AR, Newman WG, Panda B, Ramsden SC, Black GC (2017) Validation of copy number variation analysis for next-generation sequencing diagnostics. Eur J Hum Genet 25(6):719–724. https://doi.org/10.1038/ejhg.2017.42
Grasso C, Butler T, Rhodes K, Quist M, Neff TL, Moore S, Tomlins SA, Reinig E, Beadling C, Andersen M, Corless CL (2015) Assessing copy number alterations in targeted, amplicon-based next-generation sequencing data. J Mol Diagn 17(1):53–63. https://doi.org/10.1016/j.jmoldx.2014.09.008
Van Cutsem E, Bang YJ, Feng-Yi F, Xu JM, Lee KW, Jiao SC, Chong JL, Lopez-Sanchez RI, Price T, Gladkov O, Stoss O, Hill J, Ng V, Lehle M, Thomas M, Kiermaier A, Ruschoff J (2015) HER2 screening data from ToGA: targeting HER2 in gastric and gastroesophageal junction cancer. Gastric Cancer 18(3):476–484. https://doi.org/10.1007/s10120-014-0402-y
Owens MA, Horten BC, Da Silva MM (2004) HER2 amplification ratios by fluorescence in situ hybridization and correlation with immunohistochemistry in a cohort of 6556 breast cancer tissues. Clin Breast Cancer 5(1):63–69. https://doi.org/10.3816/cbc.2004.n.011
Wolff AC, Hammond MEH, Allison KH, Harvey BE, Mangu PB, Bartlett JMS, Bilous M, Ellis IO, Fitzgibbons P, Hanna W, Jenkins RB, Press MF, Spears PA, Vance GH, Viale G, McShane LM, Dowsett M (2018) Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J Clin Oncol 36(20):2105–2122. https://doi.org/10.1200/JCO.2018.77.8738
Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH, Chang MT, Chandarlapaty S, Traina TA, Paik PK, Ho AL, Hantash FM, Grupe A, Baxi SS, Callahan MK, Snyder A, Chi P, Danila D, Gounder M, Harding JJ, Hellmann MD, Iyer G, Janjigian Y, Kaley T, Levine DA, Lowery M, Omuro A, Postow MA, Rathkopf D, Shoushtari AN, Shukla N, Voss M, Paraiso E, Zehir A, Berger MF, Taylor BS, Saltz LB, Riely GJ, Ladanyi M, Hyman DM, Baselga J, Sabbatini P, Solit DB, Schultz N (2017) OncoKB: a precision oncology Knowledge Base. JCO precision oncology 2017. https://doi.org/10.1200/PO.17.00011
Bartley AN, Washington MK, Colasacco C, Ventura CB, Ismaila N, Benson AB 3rd, Carrato A, Gulley ML, Jain D, Kakar S, Mackay HJ, Streutker C, Tang L, Troxell M, Ajani JA (2017) HER2 Testing and Clinical Decision Making in Gastroesophageal Adenocarcinoma: Guideline From the College of American Pathologists, American Society for Clinical Pathology, and the American Society of Clinical Oncology. J Clin Oncol 35(4):446–464. https://doi.org/10.1200/JCO.2016.69.4836
Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M, Fitzgibbons P, Hanna W, Jenkins RB, Mangu PB, Paik S, Perez EA, Press MF, Spears PA, Vance GH, Viale G, Hayes DF, American Society of Clinical O, College of American P (2014) Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med 138(2):241–256. https://doi.org/10.5858/arpa.2013-0953-SA
Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, Schnall-Levin M, White J, Sanford EM, An P, Sun J, Juhn F, Brennan K, Iwanik K, Maillet A, Buell J, White E, Zhao M, Balasubramanian S, Terzic S, Richards T, Banning V, Garcia L, Mahoney K, Zwirko Z, Donahue A, Beltran H, Mosquera JM, Rubin MA, Dogan S, Hedvat CV, Berger MF, Pusztai L, Lechner M, Boshoff C, Jarosz M, Vietz C, Parker A, Miller VA, Ross JS, Curran J, Cronin MT, Stephens PJ, Lipson D, Yelensky R (2013) Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 31(11):1023–1031. https://doi.org/10.1038/nbt.2696
Fehm T, Muller V, Aktas B, Janni W, Schneeweiss A, Stickeler E, Lattrich C, Lohberg CR, Solomayer E, Rack B, Riethdorf S, Klein C, Schindlbeck C, Brocker K, Kasimir-Bauer S, Wallwiener D, Pantel K (2010) HER2 status of circulating tumor cells in patients with metastatic breast cancer: a prospective, multicenter trial. Breast Cancer Res Treat 124(2):403–412. https://doi.org/10.1007/s10549-010-1163-x
Gao J, Wang H, Zang W, Li B, Rao G, Li L, Yu Y, Li Z, Dong B, Lu Z, Jiang Z, Shen L (2017) Circulating tumor DNA functions as an alternative for tissue to overcome tumor heterogeneity in advanced gastric cancer. Cancer Sci 108(9):1881–1887. https://doi.org/10.1111/cas.13314
Mishima Y, Matsusaka S, Chin K, Mikuniya M, Minowa S, Takayama T, Shibata H, Kuniyoshi R, Ogura M, Terui Y, Mizunuma N, Hatake K (2017) Detection of HER2 amplification in circulating tumor cells of HER2-negative gastric Cancer patients. Target Oncol 12(3):341–351. https://doi.org/10.1007/s11523-017-0493-6
Acknowledgments
We acknowledge all other studies that support our work and were not cited due to length limitations. We would like to thank all the patients who have participated in this study. We are also thankful to the doctors from departments of oncology, doctors from departments of pathology for technics and staff of the Novogene Bioinformatics. This article has been edited by Jeremy Dean Chapnick, English Language Editor, AME Publishing Company.
Availability of Data and Materials
The datasets supporting the conclusions of this article are included within the article and additional files.
Funding
This study was funded by National Natural Science Foundation of China (81472743).
Author information
Authors and Affiliations
Contributions
Jiafu Ji, Dongmei Lin and Yang Yu conceived the research idea and designed this study. Zhongwu Li, Lixin Zhou and Ling Jia performed sample collection. Yun Gao, Donfeng Niu, Lei Li, Wanchun Zang and Lianju Gao analyzed and interpreted data. All authors participated in writing, review and final approval of the manuscript.
Corresponding authors
Ethics declarations
This study was approved by the ethics committee of Peking University Cancer Hospital (2014TW04), and written informed consent was obtained from all patients.
Competing Interests
Lei Li, Wanchun Zang, Lianju Gao, Guanhua Rao, Yun Gao, Gang Cheng and Yang Yu were employee of Novogene Bioinformatics Technology Co., Ltd., Beijing, China. No potential conflicts of interest were disclosed by the other authors. SJK has received consulting/advisory fees from Lilly Oncology, Astellas, Boston Biomedical and Foundation Medicine, Inc. SJK has stock/equity in TP Therapeutics and receives institutional research funding from Merck, Leap therapeutics, and Astellas Pharmaceuticals.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary figure 1
The summary of this study. (TIF 710 kb)
Supplementary figure 2
Distribution of HER2 copy number from 151 HER2-negative FFPE specimens using FISH. A total of 151 HER2 negative samples were used to determine a cut-off value. The distribution of copy number from 151 samples followed a normal distribution (P value of Shapiro-Wilk test = 0.0539). (TIF 306 kb)
Supplementary figure 3
FISH and NGS results of six discrepancies in breast cancer specimens. a, c, e, g, i and k showed the HER2 FISH results, whereas b, c, d, g, f, h, j and l showed corresponding NGS detection of HER2 copy number. The y axis represents the log2 copy number ratio of each amplicon from each gene. (TIF 12266 kb)
ESM 4
(DOCX 12 kb)
ESM 5
(DOCX 13 kb)
ESM 6
(DOCX 15 kb)
ESM 7
(DOCX 22 kb)
ESM 8
(DOCX 44.7 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Niu, D., Li, L., Yu, Y. et al. Evaluation of Next Generation Sequencing for Detecting HER2 Copy Number in Breast and Gastric Cancers. Pathol. Oncol. Res. 26, 2577–2585 (2020). https://doi.org/10.1007/s12253-020-00844-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-020-00844-w