Abstract
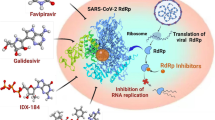
Most recently, an outbreak of severe pneumonia caused by the infection of SARS-CoV-2, a novel coronavirus first identified in Wuhan, China, imposes serious threats to public health. Upon infecting host cells, coronaviruses assemble a multi-subunit RNA-synthesis complex of viral non-structural proteins (nsp) responsible for the replication and transcription of the viral genome. Therefore, the role and inhibition of nsp12 are indispensable. A cryo-EM structure of RdRp from SARs-CoV-2 was used to identify novel drugs from Northern South African medicinal compounds database (NANPDB) by using computational virtual screening and molecular docking approaches. Considering Remdesivir as the control, 42 compounds were shortlisted to have docking score better than Remdesivir. The top 5 hits were validated by using molecular dynamics simulation approach and free energy calculations possess strong inhibitory properties than the Remdesivir. Thus, this study paved a way for designing novel drugs by decoding the architecture of an important enzyme and its inhibition with compounds from natural resources. This disclosing of necessary knowledge regarding the screening and the identification of top hits could help to design effective therapeutic candidates against the coronaviruses and design robust preventive measurements.
Graphic abstract

Similar content being viewed by others
1 Introduction
The viruses of the family Coronaviridae are now notoriously famous for their diseases causing capabilities in birds, humans and mammals. The corona virion typically composed of RNA enclosed in enveloped protein, having glycoprotein spikes, is capable of infecting a broad range of hosts, including humans. Coronaviruses, as the number of variants and diversity increases in this family, based on similarities are classified into four sub-genera, designated as alpha (α), beta (β), gamma (γ) & delta (δ) [1]. So far, the β coronaviruses (CVs) are known to cause infections in humans, including common colds and primarily affecting the respiratory system. Bats are associated with the CVs pandemics in the human population, bats harbor the virus and are believed to be immune to the viral infection itself, promoting the mutations that are crucial for the CVs pathogenicity [2]. The spike-like glycoprotein (S), giving the virus its corona like appearance is vital for their pathogenicity and helps them to attach with the host cell surface receptors and also delimits the hosts’ range for the CVs [3].
The CVs genome, ranging from 27 to 32 kilo-bases, are positive-sense single-stranded RNA (+ ssRNA) coding for, ORF1a and ORF1b, the poly-proteins involved in RNA polymerization (RNA-dependent RNA-polymerases) (RdRp) and also for modulation of host responses [4, 5]. Fatal diseases causing zoonotic strains in this family are severe acute respiratory syndrome (SARS) and the Middle East respiratory syndrome (MERS) [6]. Additionally, there are four more strains, which are reported to be disease-causing in humans, mainly common colds in individuals with immunodeficiency (229E, HKU1, NL63 and OC43) [4].
The 2019-novel-corona-virus (SARS-CoV-2) that emerged in Wuhan in 2019 belongs to a bat derived Coronaviridae family, that have gained the transmission capability from animals to humans and from human to human, due to which SARS-CoV-2 became so lethal and caused global emergency [7]. The SARS-CoV-2 is an enveloped RNA virus with the distinctive corona like shape protein spikes (usually about nine to twelve nanometers) capable of attachment to host cells. The SARS-CoV-2 potentially causes “novel corona-virus-infected pneumonia” or NCIP, the disease of lower respiratory tract having common cold-like symptoms with fever chest congestion leading to difficulty in breathing [8].
The SARS-CoV-2 has an 86.9% similarity with the genome of bat-like SARS-CVs and was classified as a distinctive subclade in the subgenus of sarbecovirus having typical β-CVs genome organization [8]. The SARS-CoV-2 genome, like other CoVs constitutes a 5ʹ untranslated region (UTR) replicase-complex Orf1a and Orf1ab followed by protein-encoding genes for the spike (S), membrane (M), envelope (E), nucleic capsid (N) and a 3ʹ UTR [9].
The non-structural proteins (nsp) from 1 to 16 of CoVs have a vital role in their replication, while the functions of certain nsps remain elusive. The structural proteins are indispensable for viral assembly and infection, while S protein for spike has distinctive variations and helps in the attachment to the host cell surface proteins [10, 11]. The M protein having transmembrane domains binds to the nucleocapsid and shaping the virion [12, 13]. The E protein is indispensable for viral pathogenesis and is responsible for virion assembly and budding [14, 15]. The N protein is comprising of two domains, having the capability of binding with virion genome and nsp-3 protein triggering replicase-transcriptase complex and viral genome encapsulation [16,17,18].
Herein, we used a multi-steps computational pipeline to identify novel compounds against the RdRp from SARs-CoV-2. Virtual screening, docking and re-docking approaches were used followed by molecular dynamics simulation and free energy calculation. Novel hits were identified, which possess better inhibitory properties than Remdesivir. This disclosing of necessary knowledge regarding the screening of natural products and the identification of top hits could help to design effective therapeutic candidates against the coronaviruses and design robust preventive measurements.
2 Material and Methods
2.1 Phylogenetic Analysis of Coronavirus:
NCBI database was used for the retrieval of Corona viruses (RdRp region) sequences. The accession no MT042778 was used as query sequence in the NCBI Blast for obtaining highly similar sequence. For the selected sequences either we can download complete sequences or only download aligned sequences using options available in NCBI. In this study we downloaded only aligned sequence to make sure to get only RdRp region. Total 110 sequences were retrieved, 107 sequences of SARS CoV-2 were placed as ingroup and remaining three of sequences Bat-CoV were used as outgroup. These sequences were aligned with the help of Clustal software [29] using pairwise multiple sequence alignment algorithm. This data matrix was used for generating trees file using Beast software [30]. Three independent Markov chain Monte Carlo analyses of 100,000,000 steps were conducted and one best tree was saved after 2000 steps. The effective sample size (ESS) of all parameters above 200 is an indication of reliable results, which is checked on Tracer ver 1.5 [31]. Trees file was uploaded to tree annotator software for obtaining a Maximum clade credibility tree with posterior probability and branch length information. The annotated tree was visualized on Figtree software [32].
2.2 Protein Structure Preparation and Active Site Identification
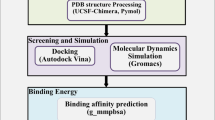
A cryo-EM structure of RdRp from SARs-CoV-2 was downloaded from RCSB using PDB ID: 6M71 [19]. The structure was subjected to energy minimization and missing residues by using the protein preparation wizard implemented in Schrodingers Maestro [20]. Structural topology was reviewed for the defects. MolProbity [21] was used to assess the quality of the constructed structure, and energy minimization was used to resolve atomic conflicts using steepest descent and gradient conjugation algorithms. The water molecules were stripped and visualized in PyMOL [22]. The active site of the RdRp is located in the seven conserved motifs from A to G. SDD sequence (residues 759, 760 and 761) K545 and R555 are reported to be a potential target for drug discovery [23]. Thus based on these residues, the active site was selected and used for virtual screening and molecular docking.
2.3 Ligands Database Retrieval, Preparation, and Virtual Screening Protocol
The database of compounds from medicinal plants from Northern south Africa was retrieved from NANPDB (https://african-compounds.org/nanpdb/) [24]. This database is a diverse source of natural drugs from 617 source species, which comes from 146 families of plants, animals, bacteria, and fungi. A SDF format file was downloaded, which comprised of 6482 compounds. Structural preparation such as charges, minimization, and compound washing was carried out. The database was then converted to.mdb format to be used as input for Molecular Operating Environment (MOE v2016) [25]. The selected residues option was used to define the active site residues. With ten conformations, each ligand was screened against the active site using a triangle matcher as a placement while London dG as a scoring method. Docking scores and visual interactions were used as a criterion for selecting the best hits.
2.4 Molecular Docking and Re-docking
Compounds obtained from virtual screening were subjected to further screening for the best active compounds against the RdRp active site. Prior to molecular docking, the obtained hits were subjected to pharmacokinetics and pharmacodynamics criteria validation, which excluded 36 compounds. The obtained 199 compounds were subjected to induced-fit docking protocol using MOE. Using the IFD protocol, the compounds were further reduced to a reasonable number, which could be then evaluated and docked individually against the active site of RdRp. For the re-docking, we used AutoDock Vina software, which is based on a Genetic Algorithm (GA) [26]. AutoDock software was used to define the grid dimension and box based on the defined residues. The protein structure was converted to .pdbqt format while using the ligands preparation criteria such as root detection, charges, hydrogen, and aromaticity criteria were used for ligands preparation. Each ligand molecule was prepared individually and converted to. pdbqt file. To achieve high accuracy, we set exhaustiveness to 64. To compare our docking results, we used Remdesivir as control. Thus here, a multi-steps docking and re-docking approaches were utilized to identify the most potential hits that could probably bypass Remdesivir in both computational and experimental setups to inhibit the SARs-CoV-2. To predict the bioactivity of each top ligand Molinspiration Cheminformatics tool was used while for ADMET analysis, SwissADME was utilized [27].
2.5 Simulation Protocol
The identified top hits and Remdesivir complexes were subjected to molecular dynamics simulation to understand the dynamics and interacting behavior of these compounds. To obtain better and accurate simulation results, all the complexes were submitted to Propka 3.1, which is an online web server, for the correction of the protonation state. Amber 18 package with pmemd.cuda implementation was used to perform the simulations [28]. The latest AMBER ff14SB force field was used for simulation. The antechamber was used to prepare the ligand topologies and obtained.frcmod file for simulation [29]. The generalized Amber force field (GAFF2) was used for small molecules parameters, and Gasteiger charges were added to each inhibitor [30].
TIP3P water box with 14 Å buffer distance each side was used to solvate the systems. Each system was neutralized by adding Na + ions. Using the 300 K temperature controlled by Langevin thermostat and a pressure of 1.0 bar scrutinized by Berendsen Barostat was used for each system [31, 32]. All bond lengths involving hydrogen atoms were constrained by the SHAKE algorithm [33]. A time step was set as 2.0 fs. For long-range interactions, particle mesh Ewald summation (PME) approach was exercised [34]. For all cases, the non-bonded cut-off was fixed at 10.0 Å. Each system was minimized by using two-step minimization approach. Followed by heating and equilibration, the production simulation was carried for 100 ns at the NPT ensemble, and the Cartesian coordinates were stored at every 10 ps. Overall, 10,000 frames were obtained from each production simulation.
2.6 Post-Simulation Analysis and Visualization
The trajectories obtained from each system were subjected to post-simulation analysis such as root mean square deviation (RMSD) to estimate the stability of each system, root mean square fluctuation (RMSF) to access the flexibility at residues level. For structure compactness, we calculated the radius of gyration as criteria for determining the structural compactness during the simulation time. For all these analyses, we used CPPTRAJ and PTRAJ [35].
2.7 Binding Free Energy Calculations
The binding of each ligand was estimated by using the molecular mechanics Poisson–Boltzmann surface area (MMGBSA) method which is a widely used and acceptable method [36,37,38,39,40]. The most widely used MMPBSA.py script was used as input, which contain all the guidelines for free energy calculations. For each system, 2500 structural frames were used to calculate the free energy using the following equation.
In this equation, ΔGbind represents total free binding energy, while others show the free energy of complex, the protein, and the ligand. Specific energy term contributes to the whole free energy was calculated by the equation:
Gbond, Gele and GvdW specify interactions among bonded, electrostatic, and van der Waals states. In contrast, Gpol and Gnpol represent the polar and non-polar interaction to the free energy presumed through precise GB (Generalized Born). This free energy calculation method is widely used by different studies to understand the binding energy of different ligands [41, 42].
3 Results
3.1 Phylogenetic Analysis
Phylogenetic analysis was conducted using molecular data (RdRp region) for the estimation of evolutionary relationship among SARS-CoV-2 members sampled from various geographical regions. In clade I the early branches are occupied by the members of USA, China and Thailand, which possibly suggest the evolution of SARS-CoV-2 in these areas. The oldest branches in clade II are occupied by the members having distribution in Thailand, which suggests that the ancestors of clade II evolved in Thailand. The members from Spain (ESP) and Jamica (JAM) occupied the early branch in clade III and IV respectively. Plesiomorphic (primitive) branches in clade V are occupied by the members of USA and Italy. This study indicates that there are two possible centers which are important in the origin and dispersal of SARs-CoV-2, these centers are East Asian center (China + Thailand) and North American center (USA). A secondary center Italy + Spain was also instrumental in proliferation of this virus.
The members of SARS-CoV-2 from Pakistan, Iran and Israil are grouped with the members from USA, whereas the Indian members are in group with Spanish and European members. Most of East Asian members are nested within Chinese members. Australian members are on same branches with Indian, East Asian and USA members (Fig. 1).
3.2 Virtual Screening and Molecular Docking
A multi-steps drug screening approach was used to search for the most potential drug candidate against the RdRp from SARs-CoV-2. A total of 6842 drugs from South African natural resources were screened in three steps. In the first step using MOE, all the compounds were screened, and the scores obtained from this screening range from − 7.0 to − 3.0 kcal/mol. To select the best compounds from all these, a criterion based on docking score and multiple interactions with the defined active site residues was used to filter the top hits. This screening resulted in 236 best compounds satisfying the specified criteria. Each conformation was manually visualized for this purpose. The obtained 236 compounds were then subjected to ADMET analysis, which excluded 37 compounds while the remaining were the best fit. Using the IFD methods, the remaining 199 compounds were again screened against the RdRp polymerase. In the case of Induced fit docking, the scores obtained were range from − 8.16 to − 4.34 kcal/mol. Here we again follow the same criteria to select the best hits using molecular docking score and visual interaction analysis. From these, 199 compounds, only 42 compounds were found to form the best interactions with active site residues and to have good binding affinity.
To further validate the activity of these final hits against the RdRp, we used the best algorithm (Genetic Algorithm) by AutoDock Vina. These 42 compounds and the receptor molecules were prepared and converted to the AutoDock Vina acceptable format (.pdbqt). Exhaustiveness was set 64 to achieve high accuracy. Results from AutoDock Vina range from − 10.4 to − 5.1 kcal/mol (Table 1). Considering Remdesivir as control, the docking score reported by AutoDock Vina was − 7.1 kcal/mol. Hence, using − 7.1 kcal/mol as a threshold, 24 compounds were found to have a docking score better than the Remdesivir docking score. Finally, these 24 compounds were analyzed, and the top 4 hits with the best docking score and Remdesivir were selected for further comparatively analysis.
3.3 Interaction Analysis of Top Hit and Remdesivir with RdRp
Analysis of the top hits and Remdesivir revealed that all the compounds possess strong inhibitory effects against the RdRp. In the case of Remdesivir, the docking score was found to be − 7.1 kcal/mol. As given in Table 2, Remdesivir forms five hydrogen bonds with the key active site residues. These residues include Lys621, Cys622, Asp761, Lys798, and Glu811. Besides, five hydrophobic and one salt bridge was also formed with different residues. On the other hand, the best compound Diosmetin-7-O-Beta-d-apiofuranoside with docking score − 10.4 kcal/mol formed nine hydrogen bonds with the key active site residues including Trp617, Tyr619, Lys621, Cys622, Asp623, Asp760, Asp761, Ala762, and Trp800. Furthermore, hydrophobic interactions with the key residues Asp618, Lys798, and Glu811 were also observed. Among the top four hits identified, the second compound 3-O-Alpha-l-arabinopyranosyl-echinocystic acid formed eight hydrogen bonds with Asp452, Thr556, Asp618, Tyr619, Lys621, Asp623, Arg624, and Asp760. Alongside salt bridges and π-Cation interactions with Arg553, Lys621, and Lys798 were also formed. The docking score for 3-O-Alpha-l-arabinopyranosyl-echinocystic acid was reported to be − 9.9 kcal/mol. Furthermore, compound 3′-epi-afroside with the docking score − 9.3 kal/mol also formed eight hydrogen bonds with Trp617, Tyr619, Lys621, Cys622, Asp623, Asp760, Asp761, Trp800 and three hydrophobic interactions with Asp618, Lys798 and Glu811 were observed. However, no salt bridge or π-Cation interaction was reported. Among the top-scoring best hits, Genkwanin 8-C-beta-glucopyranoside was also included. The docking score for the 4th ranked compound was reported to be -9.1 kcal/mol. Seven hydrogen bonds with the key active site residues such as Asp452, Arg553, Thr556, Lys621, Cys622, Asp623, Asp760, and one salt bridge with Arg555 was observed. These results are self-explanatory that the compounds identified through a multi-step screening and docking possess better inhibitory effects than Remdesivir. Not only these compounds possess the best docking scores, but also multiple interactions with the key amino acids are observed. The interaction pattern of all these compounds, including Remdesivir used as a control, are given in Fig. 2. Furthermore, details including the drug names, final docking score, interactions which includes hydrogen bonding, hydrophobic interaction, salt bridges, and π-Cation interactions are given in Table 2.
Furthermore, the bioactivity of these top hits and Remdesivir was predicted and compared. As given in Table 2, the bioactivity predicted by the Molinspiration Cheminformatics tool reported that the top four hits possess strong bioactivity against enzymes than Remdesivir. The bioactivity score for Remdesivir was reported to be 0.38, which is the same as 3-O-Alpha-l-arabinopyranosyl-echinocystic acid (0.38). The compound Diosmetin-7-O-Beta-d-apiofuranoside was reported to have the bioactivity score 0.36 while 3′-epi-afroside and Genkwanin 8-C-beta-glucopyranoside possess many fold stronger bioactivity than Remdesivir. The bioactivity scores for these two compounds were reported to be 0.75 and 0.40, respectively.
Furthermore, the ADMET properties of such as molecular weight, LogP, number of rotatable bonds, hydrogen bond donor, and acceptors were calculated for each compound. It can be seen all the four compounds obey the ADMET properties and thus increases the reliability of experimental results. All the results are given in Table 3.
3.4 Dynamics Stability and Flexibility Analysis
To understand the dynamics stability and convergence, RMSD as a function of time of all the systems was calculated. The RMSD of all five systems is given in Fig. 3. It can be scene that the Diosmetin-7-O-Beta-d-apiofuranoside complex reached a stable equilibrium after 20 ns. The system possesses stable behavior during simulation. The average RMSD for the Diosmetin-7-O-Beta-d-apiofuranoside system was observed to be 2.0 Å. On the other hand, a little convergence between 20 and 50 ns was observed in the case of 3-O-Alpha-l-arabinopyranosyl-echinocystic acid, but soon after 50 ns, the RMSD values fell, and the average RMSD was observed to be between 2.5 and 3.0 Å. Comparatively, this system remained relatively unstable than the first one. Likewise, the two other systems also remained stable during the simulation. In the case of 3′-epi-afroside, a little convergence between 60 and 70 ns was observed, but overall the system remained stable. The average RMSD for 3′-epi-afroside was decreased to be between 2.0 and 2.5 Å. However, in the case of Genkwanin 8-C-beta-glucopyranoside, the systems showed acceptable convergence at different intervals, but overall the system was stable. For Genkwanin 8-C-beta-glucopyranoside, the average RMSD was to be between 2.0 and 2.5 Å. We also simulated the Remdesivir complex to understand its behavior. In the case of Remdesivir, the average RMSD remained higher than the other. At different time intervals, acceptable convergences were observed too. Overall, these results suggest that the identified compounds possess stable behavior during the simulation.
Furthermore, we also determined residual flexibility by using Root Mean Square Fluctuation (RMSF). It is evident from Fig. 4 that all five systems display more or less similar fluctuations. In the case of Diosmetin-7-O-Beta-d-apiofuranoside and 3′-epi-afroside systems, a higher fluctuation between 50 and 80 residues can be seen while no significant differences in other regions are observed. On the other hand, Remdesivir and Genkwanin 8-C-beta-glucopyranoside posses similar fluctuation with increased fluctuation between 100–180 and 450–500 residues. In the case of Genkwanin 8-C-beta-glucopyranoside, a little higher fluctuation between 10 and 30 amino acids was observed. Furthermore, 3-O-Alpha-l-arabinopyranosyl-echinocystic acid showed a different fluctuation between 160 and 180, which is very different from others. Thus, the binding of these ligands differentially affects the internal dynamics and residual flexibility.
3.5 Radius of Gyration (Rg) Calculation
The structural compactness of each system was analyzed by estimating the radius of gyration (Rg) from their respective MD trajectories, and the average values are reported. A similar Rg is obtained for all the four systems except the Remdesivir System. The average Rg for top hits systems was found to be between 25.2 and 25.4 Å, while this value increased for Remdesivir, and the average value was reported to be 25.8 Å. Thus the four compounds (top hits) bound to RdRp imply sustained stability and compactness of the complexes. Alternatively, the higher Rg value in the case of Remdesivir than the others, causing the interactions between ligand and protein to be weaker. Thus, we speculate that these compounds explored through computational pipeline may possess robust inhibitory effects than Remdesivir in the experimental assays. All the Rg(s) calculated are given in Fig. 5.
3.6 Binding Free Energy
To estimate the binding free energy of each complex, a MM/GBSA was used. MM/GBSA is the most popular and reliable approach to calculate the binding energy of ligand during MD simulation. The total binding energy of a system calculates different energy terms such as SASA, vdW, PS, and electrostatic energy. To compare the results of the top four hits, Remdesivir, which is considered as the most potent drugs reported being active against the RdRp. The results confirmed that the four hits identified from screening, docking, and re-docking possess better binding affinities than Remdesivir. It was reported that Remdesivir possesses the total binding energy − 54.4061 kcal/mol. Whereas the other four possess − 59.486 kcal/mol (Diosmetin-7-O-Beta-d-apiofuranoside), − 57.184 kcal/mol (3-O-Alpha-l-arabinopyranosyl-echinocystic acid), − 60.315 kcal/mol (3′-epi-afroside) and − 65.695 kcal/mol (Genkwanin 8-C-beta-glucopyranoside) respectively. While the other energy terms such as van der Waals energy, electrostatic energy, polar solvation energy, solvent‐accessible surface area are given in Table 4, thus these results strongly suggest that the top hits identified here should be tested experimentally against the SARS-COV-2 at earliest.
4 Discussion
RNA-dependent RNA-polymerase is an important replicating enzyme which plays important role in the processing of RNA from SARs-CoV-2. The cry-EM structure of the RdRp recently reported revealed that the structure possesses similar architecture of Finger, Palm, Thumb and NiRAN region. A higher identity between the previously reported SAR-CoV and the recently reported structure is due to high amino acid conservancy. The study highlighted important residues, domains, and conserved motifs will help to identify potent inhibitors and help to control the emerging infections related to Coronaviridae family [19].
Computational methods are of great importance in determining the structure and function of proteins, drug binding, exploring the resistance mechanism, and bio-catalysis [41,42,43]. So, herein, using structure-based virtual screening approach shortlisted the top hits which forms important hydrogen, hydrophobic and other important interactions with the RdRp active site residues. The top hits were confirmed by performing IFD, which further shortlisted the top hits list very precisely. Using another round of docking with different algorithm exempted further hits from the list and shortlisted the top hits which could bypass Remdesivir. The use of molecular dynamics simulation technique and free energy calculations is the most widely practiced approaches while studying the protein ligand interaction. Integrating this pipeline further increased the reliability the quest to test our top hits experimentally because of its promising results. Thus, this study comprised of a complicated and multiple validations stress on the experimental assays of the top hits to help to contain the recent outbreak.
5 Conclusion
In conclusion, this study identified novel hits from natural sources. Using the structure-based approaches shortlisted the top hits which could inhibit this target experimentally. Furthermore, we also validated our shortlisted compounds by using simulation and free energy calculation. Thus, this study is a significant consideration in future strategies against the outbreaks caused by such viruses.
References
Spaan W, Cavanagh D, Horzinek M (1988) Coronaviruses: structure and genome expression. J Gen Virol 69(12):2939–2952
Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H (2005) Bats are natural reservoirs of SARS-like coronaviruses. Science 310(5748):676–679
Masters PS (2006) The molecular biology of coronaviruses. Adv Virus Res 66:193–292
Su S, Wong G, Shi W, Liu J, Lai AC, Zhou J, Liu W, Bi Y, Gao GF (2016) Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol 24(6):490–502
Khan A, Saleem S, Idrees M, Ali SS, Junaid M, Kaushik AC, Wei D-Q (2018) Allosteric ligands for the pharmacologically important Flavivirus target (NS5) from ZINC database based on pharmacophoric points, free energy calculations and dynamics correlation. J Mol Graph Model 82:37–47
Cui J, Li F, Shi Z-L (2019) Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17(3):181–192
Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 562:1–4
Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 38:1–11
Dong N, Yang X, Ye L, Chen K, Chan EW-C, Yang M, Chen S (2020) Genomic and protein structure modelling analysis depicts the origin and infectivity of 2019-nCoV, a new coronavirus which caused a pneumonia outbreak in Wuhan, China, pp 1–14
Beniac DR, Andonov A, Grudeski E, Booth TF (2006) Architecture of the SARS coronavirus prefusion spike. Nat Struct Mol Biol 13(8):751–752
Delmas B, Laude H (1990) Assembly of coronavirus spike protein into trimers and its role in epitope expression. J Virol 64(11):5367–5375
Nal B, Chan C, Kien F, Siu L, Tse J, Chu K, Kam J, Staropoli I, Crescenzo-Chaigne B, Escriou N (2005) Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J Gen Virol 86(5):1423–1434
Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, Connelly S, Droese B, Klaus JP, Makino S, Sawicki SG (2011) A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol 174(1):11–22
DeDiego ML, Álvarez E, Almazán F, Rejas MT, Lamirande E, Roberts A, Shieh W-J, Zaki SR, Subbarao K, Enjuanes L (2007) A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J Virol 81(4):1701–1713
Nieto-Torres JL, DeDiego ML, Verdia-Baguena C, Jimenez-Guardeno JM, Regla-Nava JA, Fernandez-Delgado R, Castano-Rodriguez C, Alcaraz A, Torres J, Aguilella VM (2014) Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog 10(5):1–19
Fehr AR, Perlman S (2015) Coronaviruses: an overview of their replication and pathogenesis. In: Coronaviruses. Springer, pp 1–23
Chang C-K, Sue S-C, Yu T-H, Hsieh C-M, Tsai C-K, Chiang Y-C, Lee S-J, Hsiao H-H, Wu W-J, Chang W-L (2006) Modular organization of SARS coronavirus nucleocapsid protein. J Biomed Sci 13(1):59–72
Hurst KR, Koetzner CA, Masters PS (2009) Identification of in vivo-interacting domains of the murine coronavirus nucleocapsid protein. J Virol 83(14):7221–7234
Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, Wang T, Sun Q, Ming Z, Zhang L (2020) Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 368(6492):779–782
Release S (2017) 1: Maestro. Schrödinger, LLC, New York
Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66(1):12–21
DeLano WL (2002) Pymol: an open-source molecular graphics tool. CCP4 Newslett Protein Crystallogr 40(1):82–92
Yin W, Mao C, Luan X, Shen D-D, Shen Q, Su H, Wang X, Zhou F, Zhao W, Gao M (2020) Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 368:1499–1504
Ntie-Kang F, Telukunta KK, Döring K, Simoben CV, A. Moumbock AF, Malange YI, Njume LE, Yong JN, Sippl W, Günther S (2017) NANPDB: a resource for natural products from Northern African sources. J Nat Prod 80(7):2067–2076. https://doi.org/10.1021/acs.jnatprod.7b00283
Vilar S, Cozza G, Moro S (2008) Medicinal chemistry and the molecular operating environment (MOE): application of QSAR and molecular docking to drug discovery. Curr Top Med Chem 8(18):1555–1572
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:42717
Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE III, DeBolt S, Ferguson D, Seibel G, Kollman P (1995) AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput Phys Commun 91(1–3):1–41
Wang J, Wang W, Kollman PA, Case DA (2001) Antechamber: an accessory software package for molecular mechanical calculations. J Am Chem Soc 222:U403
Vassetti D, Pagliai M, Procacci P (2019) Assessment of GAFF2 and OPLS-AA general force fields in combination with the water models TIP3P, SPCE, and OPC3 for the solvation free energy of druglike organic molecules. J Chem Theory Comput 15(3):1983–1995
Davidchack RL, Handel R, Tretyakov M (2009) Langevin thermostat for rigid body dynamics. J Chem Phys 130(23):234101
Lin Y, Pan D, Li J, Zhang L, Shao X (2017) Application of Berendsen barostat in dissipative particle dynamics for nonequilibrium dynamic simulation. J Chem Phys 146(12):124108
Kräutler V, Van Gunsteren WF, Hünenberger PH (2001) A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J Comput Chem 22(5):501–508
Toukmaji A, Paul D, John Jr A (1996) Distributed P trticle-Mesh Ewald: a Parallel Ewald Summation Method. In: PDPTA. Citeseer, pp. 33–43
Roe DR, Cheatham TE III (2013) PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput 9(7):3084–3095
Sun H, Li Y, Tian S, Xu L, Hou T (2014) Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Phys Chem Chem Phys 16(31):16719–16729
Khan A, Junaid M, Kaushik AC, Ali A, Ali SS, Mehmood A, Wei D-Q (2018) Computational identification, characterization and validation of potential antigenic peptide vaccines from hrHPVs E6 proteins using immunoinformatics and computational systems biology approaches. PLoS ONE 13(5):1–25
Junaid M, Shah M, Khan A, Li C-D, Khan MT, Kaushik AC, Ali A, Mehmood A, Nangraj AS, Choi S (2019) Structural-dynamic insights into the H. pylori cytotoxin-associated gene A (CagA) and its abrogation to interact with the tumor suppressor protein ASPP2 using decoy peptides. J Biomol Struct Dyn 37(15):4035–4050
Khan A, Junaid M, Li C-D, Saleem S, Humayun F, Shamas S, Ali SS, Babar Z, Wei D-Q (2020) Dynamics insights into the gain of flexibility by Helix-12 in ESR1 as a mechanism of resistance to drugs in breast cancer cell lines. Front Mol Biosci 6:159
Khan MT, Ali A, Wang Q, Irfan M, Khan A, Zeb MT, Zhang Y-J, Chinnasamy S, Wei D-Q (2020) Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2. A molecular dynamic study. J Biomol Struct Dyn 395:1–14
Wang Y, Khan A, Chandra Kaushik A, Junaid M, Zhang X, Wei D-Q (2019) The systematic modeling studies and free energy calculations of the phenazine compounds as anti-tuberculosis agents. J Biomol Struct Dyn 37(15):4051–4069
Khan A, Kaushik AC, Ali SS, Ahmad N, Wei D-Q (2019) Deep-learning-based target screening and similarity search for the predicted inhibitors of the pathways in Parkinson's disease. RSC Adv 9(18):10326–10339
Khan A, Muhammad J, Li C-D, Saleem S, Humayun F, Shamas S, Ali SS, Babar Z, Wei D-Q (2019) Dynamics insights into the gain of flexibility by Helix-12 in ESR1 as a mechanism of resistance to drugs in breast cancer cell lines. Front Mol Biosci 6:159
Acknowledgements
Dong-Qing Wei is supported by the grants from the Key Research Area Grant 2016YFA0501703 of the Ministry of Science and Technology of China, the National Natural Science Foundation of China (Contract No. 61832019, 61503244), the Natural Science Foundation of Henan Province (162300410060) and Joint Research Funds for Medical and Engineering and Scientific Research at Shanghai Jiao Tong University (YG2017ZD14). The computations were partially performed at the Center for High-Performance Computing, Shanghai Jiao Tong University.
Author information
Authors and Affiliations
Contributions
AK, MK, SS, ZB and SSA conceptualized the study and did the analysis. AA, AAK, FH, ZS wrote the manuscript. AK and SS revised the manuscript, performed all the additional analysis and write-up in the revised version. DQW is an academic supervisor. He supervised the study.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Khan, A., Khan, M., Saleem, S. et al. Phylogenetic Analysis and Structural Perspectives of RNA-Dependent RNA-Polymerase Inhibition from SARs-CoV-2 with Natural Products. Interdiscip Sci Comput Life Sci 12, 335–348 (2020). https://doi.org/10.1007/s12539-020-00381-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-020-00381-9