Leaf Area Index Estimation Algorithm for GF-5 Hyperspectral Data Based on Different Feature Selection and Machine Learning Methods
Abstract
:1. Introduction
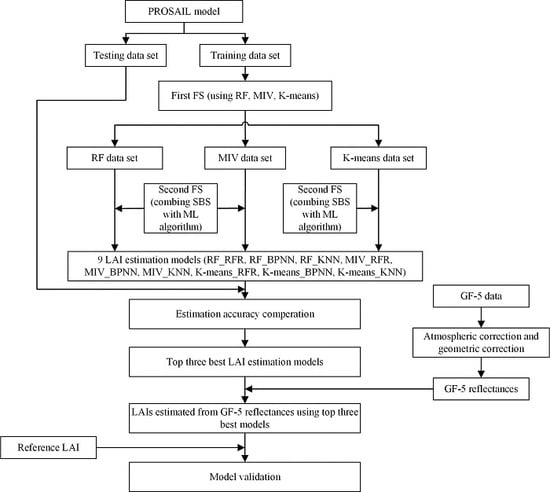
2. Materials and Methods
2.1. Study Area and Field Survey
2.2. Data Pre-Processing
2.2.1. Sentinel-2 Data
2.2.2. GF-5 Hyperspectral Data
2.3. Using PROSAIL Model to Generate Simulated Data
2.4. Feature Selection Methods
2.4.1. RF for Feature Selection
2.4.2. MIV
2.4.3. K-means
2.5. Machine Learning Algorithm
2.5.1. RFR
2.5.2. BPNN
2.5.3. KNN
2.6. LAI Estimation Accuracy Evaluation
3. Results
3.1. Determining the Dimension Number of the First FS Process
3.2. LAI Estimation Using Features Selected by the First FS Process
3.3. Optimal Bands Combination Searching in the Second FS Process
3.4. Evaluation of GF-5 LAI Estimation
4. Discussion
5. Conclusions
- (1)
- Using the same ML algorithm as feature selection and regression methods could not always ensure an optimal LAI estimation result. In this study, the RF_RFR model using the random forest algorithm as both FS and regression methods achieved higher estimation accuracy than RF_BPNN and RF_KNN when using simulated data. The MIV_BPNN is another model that uses the same algorithm as the FS and regression method. However, this model yielded lower estimation accuracy than using other regression algorithms (MIV_RFR and MIV_KNN).
- (2)
- The RF algorithm can be regarded as one of the most adaptable algorithms for further studies of biophysical parameters estimation using hyperspectral data. Not only RF-based features retained the most useful information for LAI estimation, but this algorithm was also less affected by the redundant variables when used as the regression method.
- (3)
- The proposed two-step feature selection process can achieve more satisfactory estimations with even fewer inputs. The study indicates that the feature ranking provided by RF and MIV only represents the importance of a single feature, thus the combination of high-score features could not represent the best inputs of the LAI estimation model. While the additional selection process based on the SBS algorithm was very effective in the optimal subset searching in a small or moderate dimension. Therefore, this two-step feature selection method improved the model performance by taking advantage of two FS algorithms with different criteria (first to reduce dimension, then search for the optimal subset). This proposed method was not only suitable for LAI estimation, but also can be used for classification based on hyperspectral remote sensing data.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chen, J.M.; Pavlic, G.; Brown, L.; Cihlar, J.; Leblanc, S.G.; White, H.P.; Hall, R.J.; Peddle, D.R.; King, D.J.; Trofymow, J.A.; et al. Derivation and validation of canada-wide coarse-resolution leaf area index maps using high-resolution satellite imagery and ground measurements. Remote Sens. Environ. 2002, 80, 165–184. [Google Scholar] [CrossRef]
- Karimi, S.; Sadraddini, A.A.; Nazemi, A.H.; Xu, T.; Fard, A.F. Generalizability of gene expression programming and random forest methodologies in estimating cropland and grassland leaf area index. Comput. Electron. Agric. 2018, 144, 232–240. [Google Scholar] [CrossRef]
- Wang, L.; Wang, P.; Liang, S.; Qi, X.; Li, L.; Xu, L. Monitoring maize growth conditions by training a bp neural network with remotely sensed vegetation temperature condition index and leaf area index. Comput. Electron. Agric. 2019, 160, 82–90. [Google Scholar] [CrossRef]
- Calders, K.; Origo, N.; Disney, M.; Nightingale, J.; Woodgate, W.; Armston, J.; Lewis, P. Variability and bias in active and passive ground-based measurements of effective plant, wood and leaf area index. Agric. For. Meteorol. 2018, 252, 231–240. [Google Scholar] [CrossRef]
- Hales, K.; Neelin, J.D.; Zeng, N. Sensitivity of tropical land climate to leaf area index: Role of surface conductance versus albedo. J. Clim. 2004, 17, 1459–1473. [Google Scholar] [CrossRef]
- Gonsamo, A.; Walter, J.M.; Chen, J.M.; Pellikka, P.; Schleppi, P. A robust leaf area index algorithm accounting for the expected errors in gap fraction observations. Agric. For. Meteorol. 2018, 248, 197–204. [Google Scholar] [CrossRef]
- Xiao, Z.; Liang, S.; Jiang, B. Evaluation of four long time-series global leaf area index products. Agric. For. Meteorol. 2017, 246, 218–230. [Google Scholar] [CrossRef]
- Delegido, J.; Verrelst, J.; Meza, C.M.; Rivera, J.P.; Alonso, L.; Moreno, J. A red-edge spectral index for remote sensing estimation of green LAI over agroecosystems. Eur. J. Agron. 2013, 46, 42–52. [Google Scholar] [CrossRef]
- Zhu, X.; Skidmore, A.K.; Wang, T.; Liu, J.; Darvishzadeh, R.; Shi, Y.; Premier, J.; Heurich, M. Improving leaf area index (LAI) estimation by correcting for clumping and woody effects using terrestrial laser scanning. Agric. For. Meteorol. 2018, 263, 276–286. [Google Scholar] [CrossRef]
- Soudani, K.; François, C.; Le Maire, G.; Le Dantec, V.; Dufrêne, E. Comparative analysis of IKONOS, SPOT, and ETM+ data for leaf area index estimation in temperate coniferous and deciduous forest stands. Remote Sens. Environ. 2006, 102, 161–175. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Z.; Liang, S.; Wang, J.; Chen, P. Use of general regression neural networks for generating the glass leaf area index product from time-series MODIS surface reflectance. IEEE Trans. Geosci. Remote Sens. 2014, 52, 209–223. [Google Scholar] [CrossRef]
- Huang, D.; Knyazikhin, Y.; Wang, W.; Deering, D.W.; Stenberg, P.; Shabanov, N.; Tan, B.; Myneni, R.B. Stochastic transport theory for investigating the three-dimensional canopy structure from space measurements. Remote Sens. Environ. 2008, 112, 35–50. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, P.; Bai, X.; Khan, J.; Zhang, S.; Li, L.; Wang, L. Assimilation of the leaf area index and vegetation temperature condition index for winter wheat yield estimation using Landsat imagery and the CERES-Wheat model. Agric. For. Meteorol. 2017, 246, 194–206. [Google Scholar] [CrossRef]
- Ganguly, S.; Nemani, R.R.; Zhang, G.; Hashimoto, H.; Milesi, C.; Michaelis, A.; Wang, W.; Votava, P.; Samanta, A.; Melton, F.; et al. Generating global Leaf Area Index from Landsat: Algorithm formulation and demonstration. Remote Sens. Environ. 2012, 122, 185–202. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Xiao, X.M.; Bajgain, R.; Starks, P.; Steiner, J.; Doughty, R.B.; Chang, Q. Estimating leaf area index and aboveground biomass of grazing pastures using Sentinel-1, Sentinel-2 and Landsat images. ISPRS J. Photogramm. Remote Sens. 2019, 154, 189–201. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Niu, Z.; Wang, C.; Huang, W.; Chen, H.; Gao, S.; Li, D.; Muhammad, S. Combined use of airborne Lidar and satellite GF-1 data to estimate leaf area index, height, and aboveground biomass of maize during peak growing season. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 4489–4501. [Google Scholar] [CrossRef]
- Wei, X.; Gu, X.; Meng, Q.; Yu, T.; Zhou, X.; Wei, Z.; Jia, K.; Wang, C. Leaf Area Index Estimation Using Chinese GF-1 Wide Field View Data in an Agriculture Region. Sensors 2017, 17, 1593. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.P.; Martin, R.E. Airborne spectranomics: Mapping canopy chemical and taxonomic diversity in tropical forests. Front. Ecol. Environ. 2009, 7, 269–276. [Google Scholar] [CrossRef] [Green Version]
- Thenkabail, P.S.; Smith, R.B.; Pauw, E.D. Hyperspectral vegetation indices and their relationships with agricultural crop characteristics. Remote Sens. Environ. 2000, 71, 158–182. [Google Scholar] [CrossRef]
- Perry, E.M.; Davenport, J.R. Spectral and spatial differences in response of vegetation indices to nitrogen treatments on apple. Comput. Electron. Agric. 2007, 59, 56–65. [Google Scholar] [CrossRef]
- Luo, S.; Wang, C.; Xi, X.; Pan, F.; Qian, M.; Peng, D.; Nie, S.; Qin, H.; Lin, Y. Retrieving aboveground biomass of wetland Phragmites australis (common reed) using a combination of airborne discrete-return LiDAR and hyperspectral data. Int. J. Appl. Earth Obs. Geoinf. 2017, 58, 107–117. [Google Scholar] [CrossRef]
- Ustin, S.L.; Roberts, D.A.; Gamon, J.A.; Asner, G.P.; Green, R.O. Using imaging spectroscopy to study ecosystem processes and properties. Bioscience 2004, 54, 523–534. [Google Scholar] [CrossRef]
- Smith, M.L.; Ollinger, S.V.; Martin, M.E.; Aber, J.D.; Goodale, H.C.L. Direct estimation of aboveground forest productivity through hyperspectral remote sensing of canopy nitrogen. Ecol. Appl. 2002, 12, 1286–1302. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Y.; Bao, Y.; Luo, J.; Jin, X.; Xu, X.; Song, X.; Yang, G. Exploring the best hyperspectral features for LAI estimation using partial least squares regression. Remote Sens 2014, 6, 6221–6241. [Google Scholar] [CrossRef] [Green Version]
- Meroni, M.; Colombo, R.; Panigada, C. Inversion of a radiative transfer model with hyperspectral observations for LAI mapping in poplar plantations. Remote Sens. Environ. 2004, 92, 195–206. [Google Scholar] [CrossRef]
- Zhang, X.; Liao, C.; Li, J.; Sun, Q. Fractional vegetation cover estimation in arid and semi-arid environments using hj-1 satellite hyperspectral data. Int. J. Appl. Earth Obs. Geoinf. 2013, 21, 506–512. [Google Scholar] [CrossRef]
- Jiao, Q.; Zhang, B.; Liu, J.; Liu, L. A novel two-step method for winter wheat-leaf chlorophyll content estimation using hyperspectral vegetation index. Int. J. Remote Sens. 2014, 35, 7363–7375. [Google Scholar] [CrossRef]
- Kanning, M.; Kühling, I.; Trautz, D.; Jarmer, T. High-Resolution UAV-Based Hyperspectral Imagery for LAI and Chlorophyll Estimations from Wheat for Yield Prediction. Remote Sens. 2018, 10, 2000. [Google Scholar] [CrossRef] [Green Version]
- George, R.; Padalia, H.; Sinha, S.K.; Kumar, A.S. Evaluation of the use of hyperspectral vegetation indices for estimating mangrove leaf area index in middle Andaman Island, India. Remote Sens. Lett. 2018, 9, 1099–1108. [Google Scholar] [CrossRef]
- Feng, J.; Jiao, L.; Liu, F.; Sun, T.; Zhang, X. Unsupervised feature selection based on maximum information and minimum redundancy for hyperspectral images. Pattern Recognit. 2015, 51, 295–309. [Google Scholar] [CrossRef]
- Taskin, G.; Kaya, H.; Bruzzone, L. Feature selection based on high dimensional model representation for hyperspectral images. IEEE Trans. Image Process. 2017, 26, 2918–2928. [Google Scholar] [CrossRef] [PubMed]
- Majdi, M.M.; Seyedali, M. Whale Optimization Approaches for Wrapper Feature Selection. Appl. Soft Comput. 2017, 62, 441–453. [Google Scholar]
- Samsudin, S.H.; Shafri, H.Z.M.; Hamedianfar, A.; Mansor, S. Spectral feature selection and classification of roofing materials using field spectroscopy data. J. Appl. Remote Sens. 2015, 9, 95079. [Google Scholar] [CrossRef]
- Kumar, A.; Patidar, V.; Khazanchi, D.; Saini, P. Optimizing feature selection using particle swarm optimization and utilizing ventral sides of leaves for plant leaf classification. Procedia Comput. Sci. 2016, 89, 324–332. [Google Scholar] [CrossRef] [Green Version]
- Khaled, A.Y.; Aziz, S.A.; Bejo, S.K.; Nawi, N.M.; Jamaludin, D.; Ibrahim, N.U.A. A comparative study on dimensionality reduction of dielectric spectral data for the classification of basal setm rot (BSR) disease in oil palm. Comput. Electron. Agric. 2020, 170, 105288. [Google Scholar] [CrossRef]
- Lee, J.A.; Verleysen, M. Nonlinear dimensionality reduction of data manifolds with essential loops. Neurocomputing 2005, 67, 29–53. [Google Scholar] [CrossRef]
- Alvarez-Meza, A.M.; Lee, J.A.; Verleysen, M.; Castellanos-Dominguez, G. Kernel-based dimensionality reduction using Renyi’s α-entropy measures of similarity. Neurocomputing 2017, 222, 36–46. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, X.; Li, T.; Cheng, J.; Ou, Y.; Xu, M. Dimensionality reduction of data sequences for human activity recognition. Neurocomputing 2016, 210, 294–302. [Google Scholar] [CrossRef]
- Rivera-Caicedo, J.P.; Verrelst, J.; Muñoz-Marí, J.; Camps-Valls, G.; Moreno, J. Hyperspectral dimensionality reduction for biophysical variable statistical retrieval. ISPRS J. Photogramm. Remote Sens. 2017, 132, 88–101. [Google Scholar] [CrossRef]
- Chan, K.Y.; Aydin, M.E.; Fogarty, T.C. Main effect fine-tuning of the mutation operator and the neighbourhood function for uncapacitated facility location problems. Soft Comput. 2006, 10, 1075–1090. [Google Scholar] [CrossRef]
- Imani, M.B.; Pourhabibi, T.; Keyvanpour, M.R.; Azmi, R. A new feature selection method based on ant colony and genetic algorithm on persian font recognition. Int. J. Mach. Learn. Comput. 2012, 2, 278–282. [Google Scholar] [CrossRef]
- Karegowda, A.G.; Manjunath, A.S.; Jayaram, M.A. Comparative study of attribute selection using gain ratio and correlation-based feature selection. Int. J. Inf. Technol. Knowl. Manag. 2010, 2, 271–277. [Google Scholar]
- Sylvester, E.V.; Bentzen, P.; Bradbury, I.R.; Clément, M.; Pearce, J.; Horne, J.; Beiko, R.G. Applications of random forest feature selection for fine-scale genetic population assignment. Evolut. Appl. 2018, 11, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.P.; Lin, C.J. Large-scale linear ranksvm. Neural Comput. 2014, 26, 781–817. [Google Scholar] [CrossRef] [Green Version]
- Lan, L.; Wang, Z.; Zhe, S.; Cheng, W.; Wang, J.; Zhang, K. Scaling up kernel SVM on limited resources: A low-rank linearization approach. IEEE Trans. Neural Netw. Learn. Syst. 2019, 30, 369–378. [Google Scholar] [CrossRef] [Green Version]
- Joachims, T. Making large-scale svm learning practical. Tech. Rep. 1998, 8, 499–526. [Google Scholar]
- Berger, K.; Atzberger, C.; Danner, M.; D’Urso, G.; Mauser, W.; Vuolo, F.; Hank, T. Evaluation of the PROSAIL model capabilities for future hyperspectral model environments: A review study. Remote Sens. 2018, 10, 85. [Google Scholar] [CrossRef] [Green Version]
- Darvishzadeh, R.; Skidmore, A.; Abdullah, H.; Cherenet, E.; Ali, A.; Wang, T.; Nieuwenhuis, W.; Heurich, M.; Vrieling, A.; O’Connor, B.; et al. Mapping leaf chlorophyll content from Sentinel-2 and RapidEye data in spruce stands using the invertible forest reflectance model. Int. J. Appl. Earth Obs. Geoinf. 2019, 79, 58–70. [Google Scholar] [CrossRef] [Green Version]
- Poursanidis, D.; Traganos, D.; Reinartz, P.; Chrysoulakis, N. On the use of Sentinel-2 for coastal habitat mapping and satellite-derived bathymetry estimation using downscaled coastal aerosol band. Int. J. Appl. Earth Obs. Geoinf. 2019, 80, 58–70. [Google Scholar] [CrossRef]
- Xie, Q.; Dash, J.; Huete, A.; Jiang, A.; Yin, G.; Ding, Y.; Peng, D.; Hall, C.C.; Brown, L.; Shi, Y.; et al. Retrieval of crop biophysical parameters from Sentinel-2 remote sensing imagery. Int. J. Appl. Earth Obs. Geoinf. 2019, 80, 187–195. [Google Scholar] [CrossRef]
- Pasqualotto, N.; Delegido, J.; Wittenberghe, S.A.; Rinaldi, M.; Moreno, J. Multi-Crop green LAI estimation with a new simple Sentinel-2 LAI index. Sensors 2019, 19, 904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mbulisi, S.; Onisimo, M.; Timothy, D.; Thulile, S.V.; Paramu, L.M. Estimating LAI and mapping canopy storage capacity for hydrological applications in wattle infested ecosystems using Sentinel-2 MSI derived red edge bands. Gisci. Remote Sens. 2019, 56, 68–86. [Google Scholar]
- Pearlman, J.S.; Barry, P.S.; Segal, C.C.; Shepanski, J.; Carman, S.L. Hyperion, a space-based imaging spectrometer. IEEE Trans. Geosci. Remote Sens. 2003, 41, 1160–1173. [Google Scholar] [CrossRef]
- Datt, B.; Mcvicar, T.R.; Van Niel, T.G.; Jupp, D.L.B.; Pearlman, J.S. Preprocessing eo-1 hyperion hyperspectral data to support the application of agricultural indexes. IEEE Trans. Geosci. Remote Sens. 2003, 41, 1246–1259. [Google Scholar] [CrossRef] [Green Version]
- Jacquemoud, S.; Verhoef, W.; Baret, F.; Bacour, C.; Zarco-Tejada, P.J.; Asner, G.P.; François, C.; Ustin, S.L. PROSPECT+ SAIL models: A review of use for vegetation characterization. Remote Sens. Environ. 2009, 113, 56–66. [Google Scholar] [CrossRef]
- Atzberger, C.; Richter, K. Spatially constrained inversion of radiative transfer models for improved LAI mapping from future sentinel-2 imagery. Remote Sens. Environ. 2012, 120, 208–218. [Google Scholar] [CrossRef]
- Wang, B.; Jia, K.; Liang, S.; Xie, X.; Wei, X.; Zhao, X.; Yao, Y.; Zhang, X. Assessment of Sentinel-2 MSI spectral band reflectance for estimating fractional vegetation cover. Remote Sens. 2018, 10, 1927. [Google Scholar] [CrossRef] [Green Version]
- Tao, G.; Jia, K.; Zhao, X.; Wei, X.; Xie, X.; Zhang, X.; Wang, B.; Yao, Y.; Zhang, X. Generating high spatio-temporal resolution fractional vegetation cover by fusing GF-1 WFV and MOSID data. Remote Sens. 2019, 11, 2324. [Google Scholar] [CrossRef] [Green Version]
- Weiss, M.; Baret, F.; Smith, G.J.; Jonckheere, I.; Coppin, P. Review of methods for in situ leaf area index (LAI) determination: Part II. Estimation of LAI, errors and sampling. Agric. For. Meteorol. 2004, 121, 37–53. [Google Scholar] [CrossRef]
- Baret, F.; Hagolle, O.; Geiger, B.; Bicheron, P.; Miras, B.; Huc, M.; Berthelot, B.; Niño, F.; Weiss, M.; Samain, O.; et al. Lai, fapar and fcover cyclopes global products derived from vegetation: Part 1: Principles of the algorithm. Remote Sens. Environ. 2007, 110, 275–286. [Google Scholar] [CrossRef] [Green Version]
- Ho, T.K. The random subspace method for constructing decision forests. IEEE Trans. Pattern Anal. Mach. Intell. 1998, 20, 832–844. [Google Scholar]
- Genuer, R.; Poggi, J.M.; Tuleau-Malot, C. Variable selection using random forests. Pattern Recognit. Lett. 2010, 31, 2225–2236. [Google Scholar] [CrossRef] [Green Version]
- Ou, Q.; Lei, X.; Shen, C. Individual tree diameter growth models of Larch-Spruce-Fir mixed forests based on machine learning algorithm. Forests 2019, 10, 187. [Google Scholar] [CrossRef] [Green Version]
- Rahman, M.M.; Zhang, X.; Ahmed, I.; Iqbal, Z.; Zeraatpisheh, M.; Kanzaki, M.; Xu, M. Remote sensing-based mapping of senescent leaf C:N ratio in the sundarbans reserved forest using machine learning techniques. Remote Sens. 2020, 12, 1375. [Google Scholar] [CrossRef]
- Qi, M.; Fu, Z.; Chen, F. Research on a feature selection method based on median impact value for modeling in thermal power plants. Appl. Therm. Eng. 2016, 94, 472–477. [Google Scholar] [CrossRef]
- Tan, X.; Ji, Z.; Zhang, Y. Non-invasive continuous blood pressure measurement based on mean impact value method, bp neural network, and genetic algorithm. Technol. Health Care 2018, 26, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Kanungo, T.; Mount, D.M.; Netanyahu, N.S.; Piatko, C.D.; Silverman, R.; Wu, A.Y. An efficient k-means clustering algorithm: Analysis and implementation. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 881–892. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhao, Y. The design and implementation of intrusion detection system based on data mining technology. Res. J. Appl. Sci. Eng. Technol. 2013, 5, 204–208. [Google Scholar] [CrossRef]
- Manju, V.N.; Lenin Fred, A. Ac coefficient and k-means cuckoo optimisation algorithm-based segmentation and compression of compound images. IET Image Process. 2018, 12, 218–225. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Vigneau, E.; Courcoux, P.; Symoneaux, R.; Guérin, L.; Villière, A. Random forests: A machine learning methodology to highlight the volatile organic compounds involved in olfactory perception. Food Q. Prefer. 2018, 68, 135–145. [Google Scholar] [CrossRef]
- Desai, N.P.; Lehman, C.; Munson, B.; Wilson, M. Supervised and unsupervised machine learning approaches to classifying chimpanzee vocalizations. J. Acoust. Soc. Am. 2018, 143, 1786. [Google Scholar] [CrossRef]
- Hecht-Nielsen, R. Theory of the backpropagation neural network. Neural Netw. Percept. 1992, 65–93. [Google Scholar] [CrossRef]
- Heermann, P.D.; Khazenie, N. Classification of multispectral remote sensing data using a back-propagation neural network. IEEE Trans. Geosci. Remote Sens. 1992, 30, 81–88. [Google Scholar] [CrossRef]
- Huang, R.; Xi, L.; Li, X.; Liu, C.R.; Qiu, H.; Lee, J. Residual life predictions for ball bearings based on self-organizing map and back propagation neural network methods. Mech. Syst. Signal Process. 2007, 21, 193–207. [Google Scholar] [CrossRef]
- Jia, K.; Liang, S.; Gu, X.; Baret, F.; Wei, X.; Wang, X.; Yao, Y.; Yang, L.; Li, Y. Fractional vegetation cover estimation algorithm for Chinese gf-1 wide field view data. Remote Sens. Environ. 2016, 177, 184–191. [Google Scholar] [CrossRef]
- Yang, L.; Jia, K.; Liang, S.; Liu, J.; Wang, X. Comparison of four machine learning methods for generating the GLASS fractional vegetation cover product from MODIS data. Remote Sens. 2016, 8, 682. [Google Scholar] [CrossRef] [Green Version]
- Ngia, L.S.H.; Sjoberg, J. Efficient training of neural nets for nonlinear adaptive filtering using a recursive levenberg-marquardt algorithm. IEEE Trans. Signal Process. 2000, 48, 915–1927. [Google Scholar] [CrossRef]
- Song, Y.; Liang, J.; Lu, J.; Zhao, X. An efficient instance selection algorithm for k nearest neighbor regression. Neurocomputing 2017, 251, 26–34. [Google Scholar] [CrossRef]
- Dusseux, P.; Vertès, F.; Corpetti, T.; Corgne, S.; Hubert-Moy, L. Agricultural practices in grasslands detected by spatial remote sensing. Environ. Monit. Assess. 2014, 186, 8249–8265. [Google Scholar] [CrossRef]
- Li, F.; Jin, G. Research on power energy load forecasting method based on KNN. Int. J. Ambient Energy 2019, 12, 1–7. [Google Scholar] [CrossRef]
- Gong, P.; Wang, J.; Yu, L.; Zhao, Y.; Zhao, Y.; Liang, L.; Niu, Z.; Huang, X.; Fu, H.; Liu, S. Finer resolution observation and monitoring of global land cover: First mapping results with Landsat TM and ETM+ data. Int. J. Remote Sens. 2012, 34, 2607–2654. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.S.; Cohen, W.B.; Kennedy, R.E.; Maiersperger, T.K.; Gower, S.T. Hyperspectral versus multispectral data for estimating leaf area index in four different biomes. Remote Sens. Environ. 2004, 91, 508–520. [Google Scholar] [CrossRef]
- Das, B.; Sahoo, R.N.; Pargal, S.; Krishna, G.; Verma, R.; Chinnusamy, V.; Sehgal, V.K.; Gupta, V.K. Comparative analysis of index and chemometric techniques based assessment of leaf area index (LAI) in wheat through field spectroradiometer, landsat-8, sentinel-2 and hyperion bands. Geocarto Int. 2019, 1–19. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Enclona, E.A.; Ashton, M.S.; Legg, C.; De Dieu, M.J. Hyperion, IKONOS, ALI, and ETM+ sensors in the study of African rainforests. Remote Sens. Environ. 2004, 90, 23–43. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Enclona, E.A.; Ashton, M.S.; Van Der Meer, B. Accuracy assessments of hyperspectral waveband performance for vegetation analysis applications. Remote Sens. Environ. 2004, 91, 354–376. [Google Scholar] [CrossRef]
- Bach, H.; Mauser, W. Improvements of plant parameter estimations with hyperspectral data compared to multispectral data. Proc. SPIE Int. Soc. Opt. Eng. 1997, 2959. [Google Scholar] [CrossRef]
- Mananze, S.; Pôças, I.; Cunha, M. Retrieval of maize leaf area index using hyperspectral and multispectral data. Remote Sens. 2018, 10, 1942. [Google Scholar] [CrossRef] [Green Version]
- Roberts, D.A.; Roth, K.L.; Perroy, R.L. Hyperspectral vegetation indices. In Hyperspectral Remote Sensing of Vegetation; Thenkabail, P.S., Lyon, J.G., Huete, A., Eds.; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2012; pp. 309–327. [Google Scholar]
- Halme, E.; Pellikka, P.K.E.; Mttus, M. Utility of hyperspectral compared to multispectral remote sensing data in estimating forest biomass and structure variables in Finnish boreal forest. Int. J. Appl. Earth Obs. Geoinf. 2019, 83, 101942. [Google Scholar] [CrossRef]
- Darvishzadeh, R.; Skidmore, A.; Schlerf, M.; Atzberger, C.; Corsi, F.; Cho, M.A. LAI and chlorophyll estimated for a heterogeneous grassland using hyperspectral measurements. ISPRS J. Photogramm. Remote Sens. 2008, 63, 409–426. [Google Scholar] [CrossRef]
- Darvishzadeh, R.; Atzberger, C.; Skidmore, A.K.; Abkar, A.A. Leaf area index derivation from hyperspectral vegetation indicesand the red edge position. Int. J. Remote Sens. 2009, 30, 6199–6218. [Google Scholar] [CrossRef]
- Darvishzadeh, R.; Atzberger, C.; Skidmore, A.; Schlerf, M. Mapping grassland leaf area index with airborne hyperspectral imagery: A comparison study of statistical approaches and inversion of radiative transfer models. ISPRS J. Photogramm. Remote Sens. 2011, 66, 894–906. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, K.; Liu, L.; Myint, S.W.; Wang, S.; Liu, H.; He, Z. Exploring the potential of worldview-2 red-edge band-based vegetation indices for estimation of mangrove leaf area index with machine learning algorithms. Remote Sens. 2017, 9, 1060. [Google Scholar] [CrossRef] [Green Version]
- Campos-Taberner, M.; García-Haro, F.J.; Moreno, Á.; Gilabert, M.A.; Sánchez-Ruiz, S.; Martínez, B.; Camps-Valls, G. Mapping leaf area index with a smartphone and gaussian processes. IEEE Geosci. Remote Sens. Lett. 2015, 12, 2501–2505. [Google Scholar] [CrossRef]
- Fotheringham, A.S.; Oshan, T.M. Geographically weighted regression and multicollinearity: Dispelling the myth. J. Geogr. Syst. 2016, 18, 303–329. [Google Scholar] [CrossRef]
- Dormann, C.F.; Elith, J.; Bacher, S.; Buchmann, C.; Carl, G.; Carré, G.; Marquéz, J.R.G.; Gruber, B.; Lafourcade, B.; Leitao, P.J.; et al. Collinearity: A review of methods to deal with it and a simulation study evaluating their performance. Ecography 2013, 36, 27–46. [Google Scholar] [CrossRef]
- Verrelst, J.; Rivera, J.P.; Veroustraete, F.; Muñoz-Marí, J.; Clevers, J.G.; Camps-Valls, G.; Moreno, J. Experimental sentinel-2 LAI estimation using parametric, non-parametric and physical retrieval methods—A comparison. ISPRS J. Photogramm. Remote Sens. 2015, 108, 260–272. [Google Scholar] [CrossRef]
- Siegmann, B.; Jarmer, T. Comparison of different regression models and validation techniques for the assessment of wheat leaf area index from hyperspectral data. Int. J. Remote Sens. 2015, 36, 4519–4534. [Google Scholar] [CrossRef]
- Wang, L.; Chang, Q.; Yang, J.; Zhang, X.; Li, F. Estimation of paddy rice leaf area index using machine learning methods based on hyperspectral data from multi-year experiments. PLoS ONE. 2018, 13, e0207624. [Google Scholar] [CrossRef] [Green Version]
- Verrelst, J.; Camps-Valls, G.; Muñoz Marí, J.; Rivera, J.; Veroustraete, F.; Clevers, J.; Moreno, J. Optical remote sensing and the retrieval of terrestrial vegetation bio-geophysical properties—A review. ISPRS J. Photogramm. Remote Sens. 2015, 108, 273–290. [Google Scholar] [CrossRef]
- Ge, S.; Xu, M.; Anderson, G.L.; Carruthers, R.I. Estimating yellow starthistle (centaurea solstitialis) leaf area index and aboveground biomass with the use of hyperspectral data. Weed Sci. 2007, 55, 671–678. [Google Scholar] [CrossRef]
- Liu, L.; Coops, N.C.; Aven, N.W.; Pang, Y. Mapping urban tree species using integrated airborne hyperspectral and LIDAR remote sensing data. Remote Sens. Environ. 2017, 200, 170–182. [Google Scholar] [CrossRef]
- Feilhauer, H.; Asner, G.P.; Martin, R.E. Multi-method ensemble selection of spectral bands related to leaf biochemistry. Remote Sens. Environ. 2015, 164, 57–65. [Google Scholar] [CrossRef]










| Model | Parameters | Units | Min | Max | Distribution |
|---|---|---|---|---|---|
| PROSPECT | Cab | µg/cm2 | 20 | 90 | Uniform |
| Cm | g/cm2 | 0.003 | 0.0011 | Uniform | |
| Car | µg/cm2 | 4.4 | 4.4 | - | |
| Cw | cm | 0.005 | 0.015 | Uniform | |
| Cbrown | - | 0 | 2 | Uniform | |
| Cant | µg/cm2 | 0 | 0 | - | |
| N | - | 1.2 | 2.2 | Uniform | |
| SAIL | LAI | - | 0 | 7 | Uniform |
| ALA | ° | 30 | 70 | Uniform | |
| SZA | ° | 35 | 35 | - | |
| Hot | - | 0.1 | 0.5 | Uniform |
| Model | Description |
|---|---|
| RF_RFR | Using the random forest algorithm as feature selection and regression methods. |
| RF_BPNN | Using the random forest algorithm and back propagation neural network algorithm as feature selection method and regression method, respectively. |
| RF_KNN | Using the random forest algorithm and K-nearest neighbor algorithm as the feature selection method and regression method, respectively. |
| MIV_RFR | Using the mean impact value algorithm and random forest regression algorithm as the feature selection method and regression method, respectively. |
| MIV_BPNN | Using the mean impact value algorithm and back propagation neural network algorithm as the feature selection method and regression method, respectively. |
| MIV_KNN | Using the mean impact value algorithm and K-nearest neighbor algorithm as the feature selection method and regression method, respectively. |
| K-menas_RFR | Using the K-means algorithm and random forest regression algorithm as the feature selection method and regression method, respectively. |
| K-means_BPNN | Using the K-means algorithm and back propagation neural network algorithm as the feature selection method and regression method, respectively. |
| K-means_KNN | Using the K-means algorithm and K-nearest neighbor algorithm as the feature selection method and regression method, respectively. |
| Vegetation Index | Abbreviation | Formula |
|---|---|---|
| Normalized Difference Vegetation Index | NDVI | (Band8 − Band4)/(Band8 + Band4) |
| Normalized Difference Red Edge Index 1 | NDRE1 | (Band8 − Band5)/(Band8 + Band5) |
| Normalized Difference Red Edge Index 2 | NDRE2 | (Band8 − Band6)/(Band8 + Band6) |
| Normalized Difference Red Edge Index 3 | NDRE3 | (Band8 − Band7)/(Band8 + Band7) |
| Normalized Difference Red Edge Index 4 | NDRE4 | (Band8 − Band8a)/(Band8 + Band8a) |
| FS | Machine Learning Method | Original Data Set | 20 Dimensions | ||
|---|---|---|---|---|---|
| R2 | RMSE | R2 | RMSE | ||
| RF | RFR | 0.828 | 0.837 | 0.824 | 0.849 |
| KNN | 0.764 | 0.982 | 0.768 | 0.974 | |
| BPNN | 0.797 | 0.910 | 0.791 | 0.925 | |
| MIV | RFR | 0.828 | 0.837 | 0.784 | 0.940 |
| KNN | 0.764 | 0.982 | 0.753 | 1.004 | |
| BPNN | 0.797 | 0.910 | 0.751 | 1.010 | |
| K-means | RFR | 0.828 | 0.837 | 0.819 | 0.862 |
| KNN | 0.764 | 0.982 | 0.751 | 1.008 | |
| BPNN | 0.797 | 0.910 | 0.793 | 0.921 | |
| Methods | Center Wavelength of Selected Bands of the Simulated Data and Its Corresponding Band Number of GF-5 Data |
|---|---|
| RF | 502.5 nm (A1: Band27); 527.5 nm (A2: Band33); 672.5 nm (A3: Band67); 677.5 nm (A4: Band68); 723.5 nm (A5: Band78); 728.5 nm (A6: Band80); 732.5 nm (A7: Band81); 737.5 nm (A8: Band82); 741.5 nm (A9: Band83); 1055.5 nm (A10: Band157); 1067.5 nm (A11: Band158); 1080.5 nm (A12: Band160); 1089.5 nm (A13: Band161); 1097.5 nm (A14: Band162); 1105.5 nm (A15: Band163); 1114.5 nm (A16: Band164); 1266.5 nm (A17: Band182); 2007.5 nm (A18: Band270); 2209.5 nm (A19: Band294); 2428.5 nm (A20: Band320) |
| MIV | 848.5 nm (B1: Band108); 877.5 nm (B2: Band115); 890.5 nm (B3: Band118); 937.5 nm (B4: Band129); 950.5 nm (B5: Band132); 967.5 nm (B6: Band136); 972.5 nm (B7: Band137); 1038.5 nm (B8: Band155); 1046.5 nm (B9: Band156); 1097.5 nm (B10: Band162); 1105.5 nm (B11: Band163); 1131.5 nm (B12: Band166); 1139.5 nm (B13: Band167); 1215.5 nm (B14: Band176); 1274.5 nm (B15: Band183); 1316.5 nm (B16: Band188); 1586.5 nm (B17: Band220); 1603.5 nm (B18: Band222); 1637.5 nm (B19: Band226); 1754.5 nm (B20: Band240) |
| K-means | 502.5 nm (C1: Band27); 565.5 nm (C2: Band42); 612.5 nm (C3: Band53); 668.5 nm (C4: Band66); 702.5 nm (C5: Band74); 706.5 nm (C6: Band75); 711.5 nm (C7: Band76); 723.5 nm (C8: Band79); 728.5 nm (C9: Band80); 856.5 nm (C10: Band110); 886.5 nm (C11: Band117); 997.5 nm (C12: Band143); 1080.5 nm (C13: Band160); 1148.5 nm (C14: Band168); 1494.5 nm (C15: Band209); 1519.5 nm (C16: Band212); 1754.5 nm (C17: Band240); 2007.5 nm (C18: B270); 2260.5 nm (C19: Band300); 2319.5 nm (C20: Band307) |
| FS | ML Method | R2 | RMSE | Number of Input Variables | Center Wavelength of the Selected Bands |
|---|---|---|---|---|---|
| RF | RFR | 0.828 | 0.839 | 8 | 502.5 nm; 527.5 nm; 677.5 nm; 1055.5 nm; 1080.5 nm; 1097.5 nm; 1266.5 nm; 2428.5 nm |
| BPNN | 0.794 | 0.919 | 8 | 502.5 nm; 527.5 nm; 672.5 nm; 728.5 nm; 1080.5 nm; 2007.5 nm; 2209.5 nm; 2428.5 nm | |
| KNN | 0.834 | 0.824 | 7 | 502.5 nm; 677.5 nm; 1114.5 nm; 1266.5 nm; 2007.5 nm; 2209.5 nm; 2428.5 nm | |
| K-means | RFR | 0.840 | 0.809 | 9 | 502.5 nm; 612.5 nm; 723.5 nm; 856.5 nm; 997.5 nm; 1148.5 nm; 1519.5 nm; 1754.5 nm; 2319.5 nm |
| BPNN | 0.799 | 0.906 | 9 | 502.5 nm; 565.5 nm; 668.5 nm; 702.5 nm; 723.5 nm; 856.5 nm; 1080.5 nm; 1519.5 nm; 2260.5 nm; | |
| KNN | 0.764 | 0.982 | 14 | 502.5 nm; 612.5 nm; 702.5 nm; 706.5 nm; 711.5 nm; 723.5 nm; 728.5 nm; 1080.5 nm; 1148.5 nm; 1494.5 nm; 1519.5 nm; 1754.5 nm; 2260.5 nm; 2319.5 nm | |
| MIV | RFR | 0.796 | 0.912 | 8 | 877.5 nm; 890.5 nm; 972.5 nm; 950.5 nm; 1046.5 nm; 1097.5 nm; 1215.5 nm; 1274.5 nm; |
| BPNN | 0.763 | 0.987 | 11 | 967.5 nm; 972.5 nm; 1038.5 nm; 1046.5 nm; 1097.5 nm; 1131.5 nm; 1139.5 nm; 1215.5 nm; 1274.5 nm; 1316.5 nm; 1637.5 nm; | |
| KNN | 0.777 | 0.953 | 8 | 967.5 nm; 972.5 nm; 1097.5 nm; 1105.5 nm; 1139.5 nm; 1215.5 nm; 1274.5 nm; 1316.5 nm |
| Data Set | Simulated Data | GF-5 Data | Optimal Number of Components | ||
|---|---|---|---|---|---|
| R2 | RMSE | R2 | RMSE | ||
| RF-based (20 bands) | 0.822 | 0.841 | 0.547 | 1.168 | 3 |
| MIV-based (20 bands) | 0.791 | 0.918 | 0.523 | 1.346 | 2 |
| K-means-based (20 bands) | 0.809 | 0.922 | 0.528 | 1.199 | 3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Jia, K.; Xiao, C.; Wei, D.; Zhao, X.; Lan, J.; Wei, X.; Yao, Y.; Wang, B.; Sun, Y.; et al. Leaf Area Index Estimation Algorithm for GF-5 Hyperspectral Data Based on Different Feature Selection and Machine Learning Methods. Remote Sens. 2020, 12, 2110. https://doi.org/10.3390/rs12132110
Chen Z, Jia K, Xiao C, Wei D, Zhao X, Lan J, Wei X, Yao Y, Wang B, Sun Y, et al. Leaf Area Index Estimation Algorithm for GF-5 Hyperspectral Data Based on Different Feature Selection and Machine Learning Methods. Remote Sensing. 2020; 12(13):2110. https://doi.org/10.3390/rs12132110
Chicago/Turabian StyleChen, Zhulin, Kun Jia, Chenchao Xiao, Dandan Wei, Xiang Zhao, Jinhui Lan, Xiangqin Wei, Yunjun Yao, Bing Wang, Yuan Sun, and et al. 2020. "Leaf Area Index Estimation Algorithm for GF-5 Hyperspectral Data Based on Different Feature Selection and Machine Learning Methods" Remote Sensing 12, no. 13: 2110. https://doi.org/10.3390/rs12132110