Wnt Genes in Wing Pattern Development of Coliadinae Butterflies
- 1Department of Biological Sciences, College of Sciences and Mathematics, Auburn University, Auburn, AL, United States
- 2Department of Biological Sciences, Mississippi State University, Starkville, MS, United States
- 3Division of Biological Sciences, College of Humanities and Sciences, University of Montana, Missoula, MT, United States
- 4Department of Biological Sciences, Columbian College of Arts and Sciences, The George Washington University, Washington, DC, United States
- 5Department of Biology, College of Natural Sciences, University of Puerto Rico—Rio Piedras, San Juan, Puerto Rico
- 6Molecular Sciences and Research Center, University of Puerto Rico, San Juan, Puerto Rico
- 7Department of Biochemistry, Molecular Biology, Entomology, and Plant Pathology, College of Agriculture and Life Sciences, Mississippi State University, Starkville, MS, United States
Wnt genes are major developmental genes highly conserved across all animals. Yet, our understanding of the Wnt gene repertoire and their functions is still largely incomplete. In Lepidoptera, Wnt genes have been implicated in wing pattern development. For example, WntA has been shown as a driver of wing pattern diversification in nymphalid butterflies. In this study, we characterize the Wnt gene repertoire in Zerene cesonia (Family: Pieridae), which diverged from nymphalids ∼51 million years ago, to determine if Wnt genes may have a conserved role in wing development across distant butterfly lineages. We first show that Wnt gene content is highly conserved across butterflies, but that there is strikingly different expression across the eight Wnt genes during wing development of Z. cesonia and nymphalid butterflies. Surprisingly, while four Wnt genes (Wnt1, 6, 7, and 11) are expressed during wing development in Z. cesonia, the renown nymphalid wing patterning gene WntA was undetected. However, despite the differences in Wnt gene expression, Heparin injections yielded a similar disruption in wing color pattern development in both Z. cesonia and nymphalid butterflies. Interestingly, these assays also resulted in the loss of structurally based ultraviolet (UV) wing patterns in Z. cesonia. Collectively, the study shows that pierids maintained the highly conserved nature of Wnt gene content in Lepidoptera, but might deploy these genes very differently from nymphalid butterflies during wing pattern development.
Introduction
Wnt genes are responsible for a variety of vital developmental processes and are highly conserved across animals. From axis formation in embryogenesis to insect wing development, Wnt genes are a major part of the developmental toolkit. There are 13 subfamilies of Wnt ligand genes that code for a set of secreted glycoproteins rich in cysteine. The transduction of these Wnt ligands is controlled by the canonical Wnt/β-catenin pathway and the non-canonical Wnt/JNK planar cell polarity (PCP) and Wnt/Ca2+ pathways (Rao and Kühl, 2010). The generalized function of Wnt genes in these pathways has been studied in detail over the past century (Nusse, 2012); however, the explicit function and contribution of each Wnt gene throughout development remains poorly understood, and most of what is known comes from model organisms such as mice and Drosophila. Attempts to extrapolate Wnt gene function from one model organism to another species have proven difficult, not only due to differences in the Wnt gene repertoires among species, but also the potential functions of different Wnt genes interacting in multiple Wnt pathways (Murat et al., 2010; Range, 2018). To better understand the role of Wnt ligands in the evolution of developmental processes, a systematic analysis of Wnt gene content and activity is needed in clades where Wnt ligands may have played an important role in morphological diversification.
Studies of the characterization and function of Wnt signaling among arthropods are limited outside of Drosophila. Among arthropod species that have been examined, the numbers of Wnt gene content vary from six in Acyrthosiphon pisum to nine in Tribolium castaneum (Ding et al., 2019). Among holometabolous insects, there are eight Wnt genes that are most commonly found: Wnt1, Wnt5, Wnt6, Wnt7, Wnt9, Wnt10, Wnt11, and WntA. Drosophila lacks only WntA; the functions of its seven Wnt genes include axis patterning in embryogenesis, control of the sexual dimorphic development of reproductive organs, development of neuroblast identity, and wing formation (Murat et al., 2010). Lepidoptera typically harbor a repertoire of eight Wnt genes, Wnt1, Wnt5, Wnt6, Wnt7, Wnt9, Wnt10, Wnt11, and WntA that are expressed in various tissues and stages (Ding et al., 2019; Holzem et al., 2019). In all arthropods (Murat et al., 2010) and in other animal phyla (Somorjai et al., 2018) Wnt9–Wnt1–Wnt6–Wnt10 are arranged in a tandem array, forming a syntenic block of paralogs. Not only are these genes linked on a single chromosome, but some (Wnt1–Wnt6–Wnt10) also share similarities in expression patterns on the wing discs of wing-spotted fruit flies, Drosophila guttifera (Koshikawa et al., 2015) and the Euphydryas chalcedon butterfly (Martin and Reed, 2014). Such consistent expression patterns between diptera and lepidoptera suggest a possible ancestral role of these Wnts in wing development. In addition, Wnt7–Wnt5–WntA form a syntenic arrangement of genes that is conserved across insects (Murat et al., 2010), and seem to originate from an ancestral Wnt5-7 cluster that pre existed at the root of Bilateria (Somorjai et al., 2018). Finally, Wnt11 is not located within either lepidopteran Wnt cluster, but is on another chromosome separate from the two main clusters (Ding et al., 2019).
Recently, Wnt genes have been implicated in the development and evolution of color patterns on butterfly wings. Early studies in Heliconius butterflies used genetic mapping, gene expression assays, and pharmacological manipulation to identify WntA as the gene responsible for major differences in melanic forewing patterns across the genus (Martin et al., 2012). Recently, the functional role of WntA in wing pattern development was shown in twelve Heliconius species using CRISPR gene-editing (Concha et al., 2019). Further, the role of WntA in wing pattern development has also been explored across other nymphalid butterflies, which all showed unique changes in wing pattern, including the white vein contour markings of Monarch wings (Gallant et al., 2014; Martin and Reed, 2014; Mazo-Vargas et al., 2017). Beyond this work in nymphalid butterflies, little is known about the importance of the Wnt landscape in various aspects of insect development (Murat et al., 2010).
Within Lepidoptera, the most well-characterized Wnt gene beside WntA, is Wnt1, for its role in color pattern development. Wnt1, designated as wingless when it was first discovered in Drosophila, is responsible for the development of imaginal discs, and is therefore vital for the formation of both wings and eyes (Baker, 1988). Wnt1 is perhaps most extensively studied of the Wnt genes due to its role in segment polarity (Baker, 1987). In the silkworm, Bombyx mori, Wnt1 has likely retained this ancestral role, as Wnt1 knockouts with CRISPR/Cas9 reveal its necessity during embryogenesis (Edayoshi et al., 2015; Zhang et al., 2015). In nymphalid butterflies, Wnt1 retains its expected embryonic expression (Holzem et al., 2019) and is also expressed in the peripheral tissue of the developing wings (Macdonald et al., 2010), as in other holometabolous insects (Tomoyasu et al., 2009). Within those developing wing tissues, butterfly Wnt1 also pre-patterns the “Discalis” ground plan elements on the wing across several lepidopteran lineages (Martin and Reed, 2010). In at least one nymphalid butterfly, Wnt6 and Wnt10 wing disc expression is concurrent with Wnt1 in both the peripheral tissue and discal pattern elements of the larval wing disk (Martin and Reed, 2014); an overlap that is consistent with the previously described functional redundancy between these three paralogs in Drosophila and Tribolium (Bolognesi et al., 2008; Murat et al., 2010). Quantitative PCR profiles of the silkworm Wnt repertoire also suggest that Wnt1, Wnt6, Wnt7, Wnt10, and WntA are all expressed in larval wing discs (Ding et al., 2019). Similarly, an RNA-seq study of pupal wing development in passionvine butterflies revealed expression of Wnt1, Wnt5, Wnt6, Wnt7, and WntA (Hanly et al., 2019). To our knowledge, Wnt9 and Wnt11 have not been shown to be expressed during lepidopteran wing development, and Wnt9 (syn. DWnt4) has been implicated in differences in male and female reproductive development in Drosophila (Cohen et al., 2002; Murat et al., 2010).
Here, we characterize the repertoire of Wnt genes in a pierid butterfly, Zerene cesonia, and explore their expression and potential role during wing pattern development. Z. cesonia is a Coliadinae butterfly, which are characterized by their strikingly bright yellow wings that contrast sharply with black margins often found on the wing edge. They are also noted for the bright ultraviolet (UV) reflectance of male wings, produced by specialized nano-structures on yellow pterin scales in the medial region of the forewing (Fenner et al., 2019). Among Colidinae species, UV brightness is the top indicator of male mating success (Papke et al., 2007) and we are interested in whether Wnt genes may play similar roles in pattern development in Z. cesonia as to what has been observed in other butterflies.
Pierid butterflies diverged from better studied Nymphalidae, which includes Monarchs and Heliconius butterflies, ∼51 million years ago, and from silkmoths > 95 million years ago, providing a comparative framework in Lepidoptera to explore Wnt evolution and function in wing development (Kawahara et al., 2019). Using an RNA-seq approach, we detect the expression of four out of eight Wnt genes during the wing development of Z. cesonia. Interestingly, we find different Wnt genes are expressed in Z. cesonia relative to nymphalid wings, including no expression of WntA, despite its involvement in wing pattern development of several nymphalid species. Using pharmaceutical assays similar to studies of nymphalids, we observed a similar expansion of melanic scales across the forewing in two species of pierid butterflies, Z. cesonia and its close relative Colias eurytheme. Importantly, we also observed a loss of the UV reflective pattern in Z. cesonia. Our findings suggest differences in the way the Wnt repertoire is used for color patterning during the wing development of pierids and nymphalids.
Materials and Methods
Gene Expression Assays
RNA was extracted using TRIZOL from developing wing tissue of male Z. cesonia from fifth instar larvae and every day of pupation till emergence (i.e., days 1–6). A minimum of three males were used for each stage (with a maximum of five replicates on day 4 of pupation). Total RNA was extracted and mRNA was isolated using oligo(dT) magnetic beads from NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB, Ipswich, MA, United States). cDNA was prepared using a modified version of the Smart-seq2 protocol (Picelli et al., 2014) and libraries were prepared using a Nextera XT DNA Library Preparation Kit (Illumina, San Diego, CA, United States). Libraries were sequenced as 150 base pair paired-end reads, at an average of 18.9 million reads per library (minimum of 13.2 million reads and maximum of 28.19 million reads). RNA-seq reads were preprocessed using AfterQC (Chen et al., 2017) and Trimmomatic (Bolger et al., 2014). Reads were then mapped to gene models from the reference genome using Bowtie2 and gene expression was estimated using RSEM (Li and Dewey, 2011) with default parameters. Wnt expression profiles were identified for Wnt1/wingless, Wnt6, Wnt7, and Wnt11, but no RNA-seq reads mapped to WntA, Wnt10, Wnt5, or Wnt9. Expression profiles are reported as mean FPKM (fragments per kilobase of transcript per million mapped reads) values across individuals for each developmental stage.
Gene Content Analyses
We used a combination of bioinformatic approaches to identify the repertoire of Wnt genes in representative Lepidoptera, using the Drosophila melanogaster wnt repertoire as a reference. We first surveyed the Ensembl Metazoa database (Howe et al., 2020) for the full set of wnt paralogs in B. mori, Danaus plexippus, Heliconius melpomene, and Melitaea cinxia. We then used BLAST (Altschul et al., 1990) to search for the Wnt paralogs in Bicyclus anynana, Pieris rapae, and Vanessa tameamea, which are available at NCBI. Wnt genes for Pieris napi and Phoebis sennnae were acquired using BLAST tools and available genome assemblies at lepbase.org (Challi et al., 2016). Wnt genes were identified in Z. cesonia using the annotated genome assembly v.1.0 (NCBI Accession JAAMXH000000000) (Rodriguez-Caro et al., 2020) and confirmed through sequence similarity searches and alignments with BLAST and MUMmer3.23 (Altschul et al., 1990; Kurtz et al., 2004). A Zerene eurydice, the sister species to Z. cesonia, individual (collected in San Bernardino, CA, United States) was sequenced (150 bp paired end Illumina reads, 6 Gb data yielding 20X coverage) and reads were mapped to the Z. cesonia genome assembly v1.0. Z. eurydice Wnt gene sequences were extracted using the Z. eurydice genotypes. For C. eurytheme, a 10X Genomics linked-read library was constructed for a single individual (collected in Osborne, MS, United States) according to protocol (10X Genomics, San Francisco, CA, United States) and sequenced by Novogene yielding 10.6 Gb of sequence data, which was assembled with Supernova (Weisenfeld et al., 2017). Assembly scripts and information are available though a github repository1. To identify the Wnt genes sequences in C. eurytheme, BLAST and MUMmer were used to search and align Z. cesonia Wnt gene sequences with the 10X assembly.
Phylogenetic Analyses
We aligned the Wnt paralogs for silkmoth (B. mori), four species of nymphalids (B. anynana, H. melpomene, Junonia coenia, V. tameamea), and five species of pierid butterflies (C. eurytheme, Phoebis sennae, P. napi, P. rapae, Z. cesonia, and Z. eurydice) based on their conceptual amino acid translation using MUSCLE (Edgar, 2004) as implemented in MEGAX (Kumar et al., 2018). We then reconstructed phylogenetic relationships among lepidopteran Wnt genes to resolve orthology. We inferred phylogenies in a maximum likelihood framework using IQ-Tree ver 1.6.6 (Nguyen et al., 2015) in the implementation available from the IQ-Tree web server (Trifinopoulos et al., 2016) last accessed on December 2019, selecting the best-fitting model of substitution identified with the ModelFinder subroutine from IQ-Tree (Kalyaanamoorthy et al., 2017). Support for the nodes was evaluated with the Shimodaira–Hasegawa approximate likelihood-ratio test (SH-aLRT), the aBayes test (Anisimova et al., 2011) and 1000 pseudoreplicates of the ultrafast bootstrap procedure (Hoang et al., 2018). We then removed the D. melanogaster sequences and used the resulting maximum likelihood tree to estimate differences in selective pressure among the lepidopteran wnt paralogs as evidenced by differences in the ratio of the rate of non-synonymous substitution per non-synonymous site (dN) to the rate of synonymous substitution per synonymous site (dS) using the codon-based maximum likelihood approach proposed by Goldman and Yang (1994) as implemented in PAML ver 4.9j (Yang, 2007). We explored variation in the dN/dS ratio among the different lepidopteran wnt paralogs using the branch models implemented in the program codeml from PAML.
Pharmaceutical Assays
Zerene cesonia females were collected from a local Mississippi prairie (33°30′36.98″N, 88°44′14.57″W). Females were allowed to oviposit eggs on Dalea purpurea. Upon hatching, larvae were raised on an artificial diet (Shelby et al., 2020) under 16:8 photoperiod and 28°C conditions. A laboratory colony was then maintained over several generations under these conditions. At 12 h after pupation, Z. cesonia individuals (N = 25) were injected with either 0.5 μg of heparin sodium salt (Sigma–Aldrich) diluted in water, or with water as a control, and C. eurytheme (N = 5) were injected with 0.25 μg of heparin. Injections were conducted with a 10 μl Hamilton syringe with a 26-gauge needle into the right side in the interstice that separates the baso-posterior sections of the developing wings. The outer pupal casing was sterilized with ethanol prior to injections and the pupa were hung in a pupal chamber until emergence. Individuals were preserved for photography and microscopy following eclosion.
Photography and Microscopy
Individuals were photographed using an UV-IR converted Nikon D7000 camera with an AF-S Micro Nikkor 105 mm Lens for visual color images. UV color images used the same lens and camera but with the addition of a BAADER U-Filter, 60 nm IR, and VIS filter. Scanning electron microscopy was performed on a JEOL JSM-6500F FE-SEM at 5 kV. Wing sections of Z. cesonia and specimens of Heliconius sara from Martin et al. (2012) were mounted on aluminum stubs with silver paste and coated with 15 nm of platinum.
Results
Wnt Gene Repertoire and Evolution in Butterflies
The Wnt gene content is conserved across all Lepidopteran genomes studied. Our phylogenetic analyses grouped the Lepidopteran Wnt paralogs into eight strongly supported clades that correspond to previously described Wnt genes in nymphalid butterflies (Wnt1/wingless, Wnt5, Wnt6, Wnt7, Wnt9, Wnt10, Wnt11, and WntA). In the Z. cesonia assembly v1.0, the eight Wnt genes are distributed in clusters on three chromosomes: Wnt 9-1-6-10 on chromosome 1, Wnt A-5-7 on chromosome 25, and Wnt11 on chromosome 11 (Figure 1 and Supplementary Table S1). This was not the case for Wnt11 and WntA, which have been secondarily lost in the genome of D. melanogaster. The D. melanogaster Wnt7 paralog was placed sister to the clade that included the lepidoptera Wnt11 and Wnt7 paralogs, likely as a result of deep divergence among these Wnt paralogs.
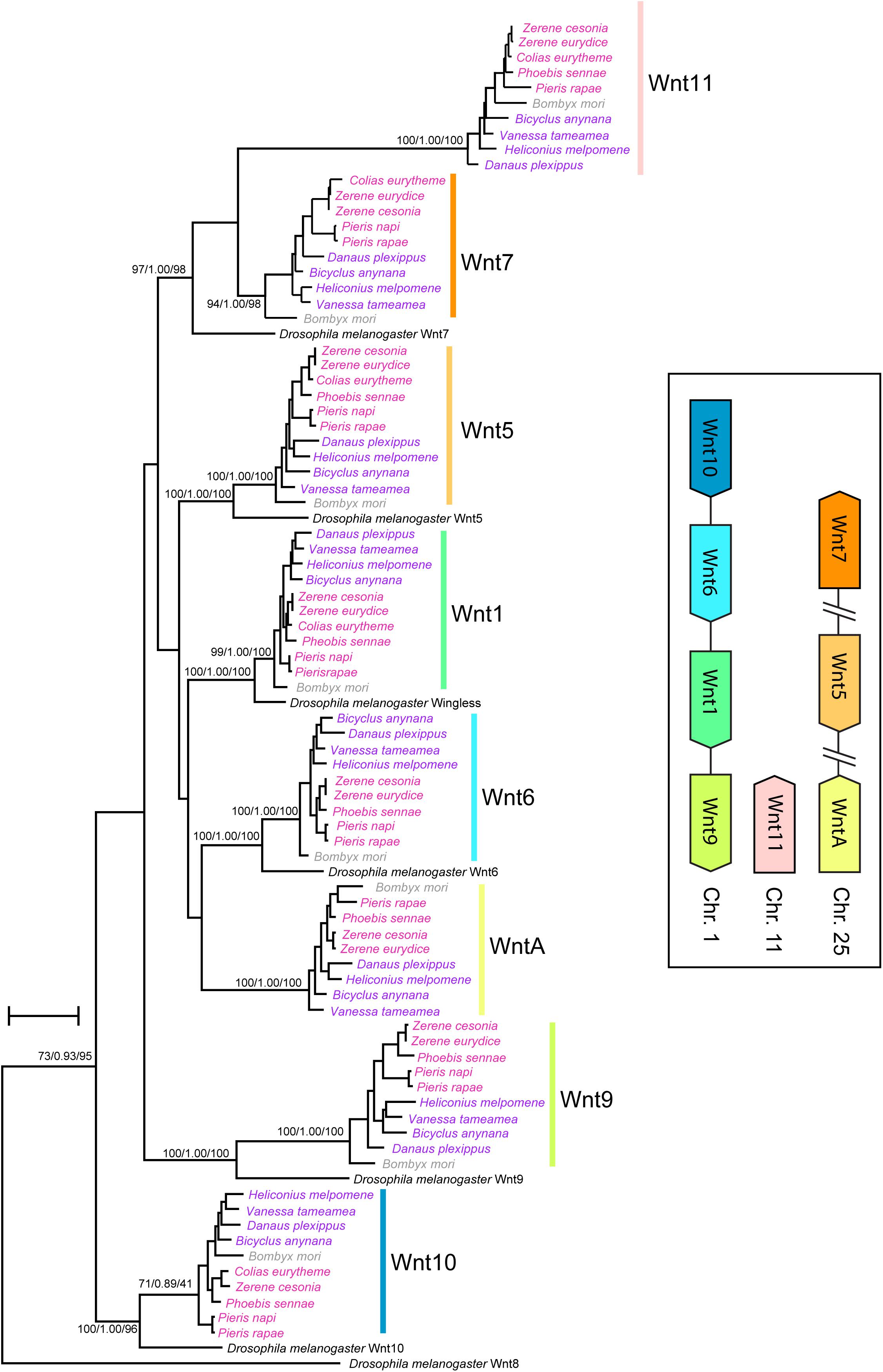
Figure 1. Wnt gene content and synteny in butterfly genomes. Phylogenetic analyses reconstruct clades for each of eight Wnt genes present in lepidoptera genomes. Inset shows chromosomal linkage and order of Wnt genes in Z. cesonia genome assembly (Supplementary Table S1), which is known to vary within clusters across Lepidoptera (Holzem et al., 2019).
Our analyses of patterns of molecular evolution failed to find significant differences in estimates of synonymous and non-synonymous substitution rates (ω) among the different lepidoptera paralogs. The model that assigns a single estimate of ω to all codons in the alignment along all branches in the tree could not be rejected in any of our tests (Supplementary Table S2), with very low value for this parameter (ω < 0.01). These findings suggest Wnt genes are evolving very slowly in Lepidoptera and are likely under stringent purifying selection due to their highly important and conserved functions during organismal development.
Wnt Gene Expression During Wing Development
Expression of Wnt genes during wing development of Z. cesonia was similar and divergent from patterns described in other lepidopteran species. Fifth instar larval wing disks showed strong expression of Wnt1 and Wnt6, consistent with their known expression in lepidoptera wing peripheral tissues (Macdonald et al., 2010; Martin and Reed, 2010, 2014) as well as in the Drosophila wing disk dorso-ventral boundary (Janson et al., 2001). In Z. cesonia, both genes showed decreased, but detectable levels during the pupal stages, which may be related to color patterning in the first 2 days after puparium formation (APF). Wnt10 is co-expressed with Wnt1 and Wnt6 in D. guttifera and in the nymphalid E. chalcedon (Martin and Reed, 2014; Koshikawa et al., 2015), but was not recovered in our pierid dataset. Wnt7 was expressed in both the larval stage and the first pupal day, as previously reported in two Heliconius species (Hanly, 2017). Following this initial peak, expression sharply decreased peaked again later during pupal development starting at the day 2 stage before rising again shortly at 5–6 days APF. Wnt11 has been associated with Wnt/JNK PCP pathway in mammals (Uysal-Onganer and Kypta, 2012), and its expression was detected during mid-pupal stages when scale cytoskeletal structures are forming. However, Wnt11 transcript abundance was at least twofold lower than any other Wnt gene suggesting expression may be very spatially and/or time restricted during wing development (Figure 2). Most noteworthy, is the lack of WntA expression, which is involved in wing pattern development among all the nymphalid butterflies assessed so far (Mazo-Vargas et al., 2017). For those Wnt genes not detected from the wing RNA-seq data, we also surveyed RNA-seq data from adult head and thorax previously generated to help annotate the Z. cesonia genome, and we were able to confirm expression of Wnt5, Wnt10, and WntA, but not Wnt9, suggesting the expression of Wnt9 is restricted to specific tissues and stages as hinted from other studies (Bolognesi et al., 2008; Murat et al., 2010; Chen et al., 2016).
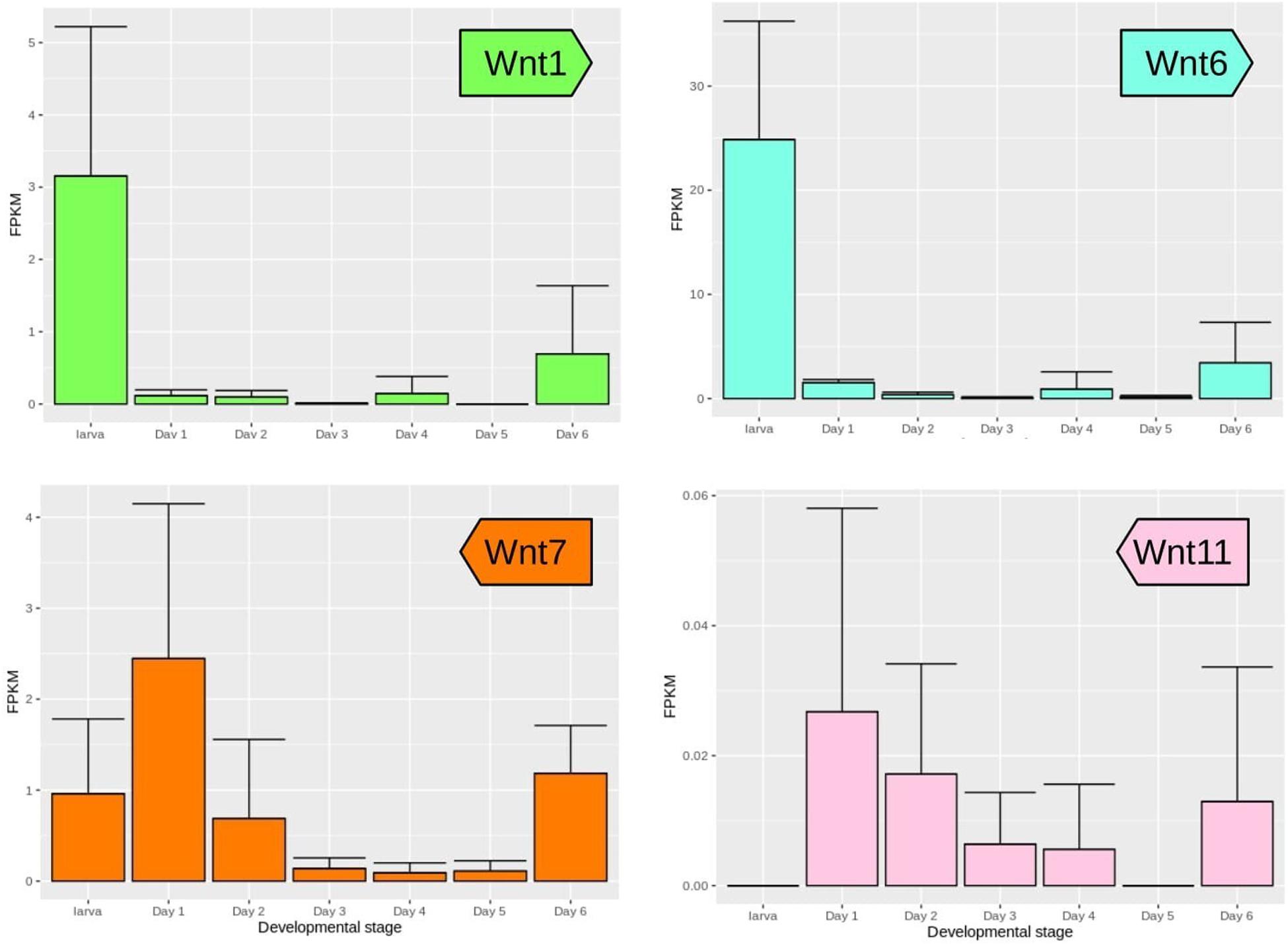
Figure 2. Wnt gene expression during wing development of Z. cesonia. Mean and standard error of expression levels among replicates from larval fifth instar through pupal wing development. Note the scale of FPKM (fragments per kilobase of transcript per million mapped reads) on y-axes differ among genes, due to large differences in expression levels.
Pharmaceutical Assays Implicate Morphogens in Wing Pattern Development
Early studies using the pharmaceutical Heparin, which can bind to Wnt ligands and facilitate their secretion and transport, induced a possible Wnt gain-of-function effect in butterfly wing patterns. These Heparin injection individuals expressed phenotypes that exhibited an extension of the basal, discal, and external symmetry systems (Serfas and Carroll, 2005). Pupal injection of Heparin in coliadinae butterflies produced melanic expansions that were reminiscent of the effects of Heparin in other butterflies (Serfas and Carroll, 2005; Martin et al., 2012; Sourakov, 2018). Heparin caused the apical melanic wing region to expand broadly across dorsal forewings in Z. cesonia females. In males, dorsal forewings showed only moderate levels of melanization but underwent instead a complete loss of the UV patterns. The dorsal hindwing melanic border was expanded in females, but unaffected in males. Both males and females exhibited ectopic expression of melanin patterning on the ventral fore and hindwings, with particularly strong melanic coloration on the apical region of the forewing. A similar trend of forewing melanin expansion was observed in C. eurytheme Heparin injected female individuals (Figure 3). Only a single male C. eurytheme injected with Heparin survived, but did not eclose properly, therefore we were unable to assay Heparin’s impact on UV reflectance in Colias. These experiments show that Heparin will perturb both pigment and structural coloration in Coliadinae butterflies in a manner similar to nymphalids, despite the lack of WntA expression. They also reveal sexually dimorphic effects of heparin that were unexpected. It is plausible that those differences highlight intrinsic divergence in the female and male patterning architecture, a hypothesis that will require further investigation but establish Z. cesonia as an interesting model system for developmental studies of sex-specific color patterning.
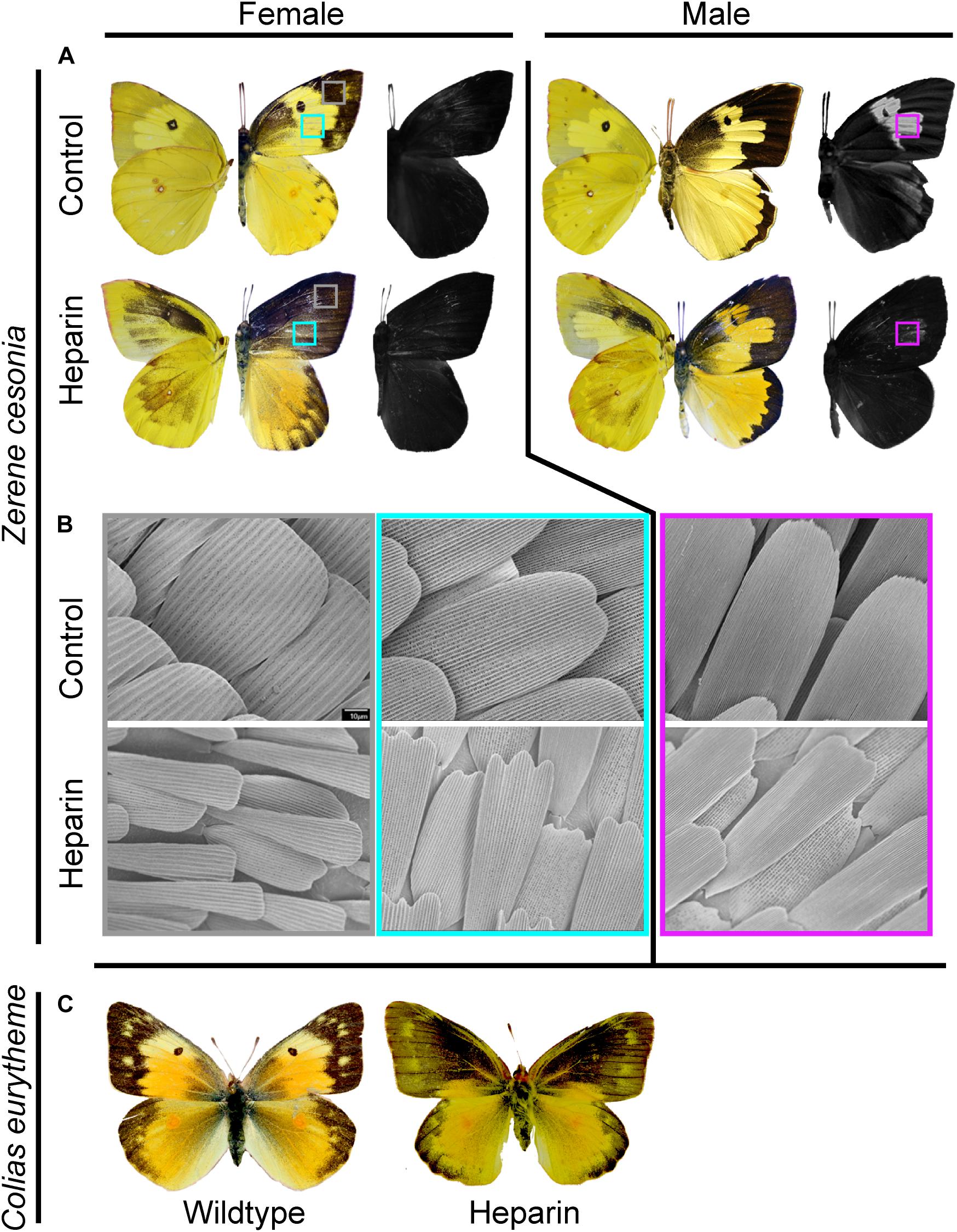
Figure 3. Heparin impacts pigment and structurally based colors on Z. cesonia wings. (A) Heparin treated female Z. cesonia showed an increase in melanization across wings and Heparin treated males lost UV reflectance. Left side wings show ventral side and right side wings show dorsal. (B) SEM imaging (1000X) of scales from control and Heparin treated wings shows difference in scale morphology and organization. (C) Female C. eurytheme dorsal wings showed melanic expansions with Heparin treatments.
Inspection of scales on Z. cesonia wings revealed broad organizational and scale shape changes (Figure 3). In the expanded melanic region of Heparin injected individuals, the melanic scales had their characteristic shapes and structures, in place of yellow scales that would typically be present. Heparin injected males that lost UV patterning did develop cover scales that appear to have the tightly organized and stacked lamellae ridges responsible for reflecting UV. However, these scales had highly reduced widths, exposing much of the ground scales that lack the UV-reflecting structures. The result was a weakly reflective UV patch on the forewings of Heparin injected male Z. cesonia (Figure 3). Scale size was highly reduced among Heparin treated individuals across multiple wing regions and both sexes (Figure 3B), suggesting that the Heparin had a systemic impact on scale development across the wings. This microscopic inspection of the impacts of Heparin demonstrates clear changes in scale structure that had not been previously documented in nymphalids.
Microscopic examination of Heparin treated Heliconius butterflies (Family: Nymphalidae) did not reveal the systemic scale reduction observed across the wings of Z. cesonia, suggesting that Heparin may affect scale development differently in the two families of butterflies. In H. sara, Heparin induced changes in both pigment and scale structure across the wing, with a complete loss of yellow scales. The black and yellow scales of H. sara are distinguishable by the shape and organization of their lamellar ridges, as seen at 5000X in Figure 4. SEM imaging revealed that Heparin induced melanic scales (Figure 4) had the expected ridge architecture of wildtype melanic scales. Similar changes in both pigment and scale structure were also observed in the CRISPR-edits of WntA among Heliconius butterflies (Concha et al., 2019), although the mutant color patterns typically altered in more narrow wing regions, since heparin is thought to also affect other Wnt ligands (Martin and Reed, 2014; Mazo-Vargas et al., 2017). In Z. cesonia, we did not see a concordant change in pigment and structure across the wing. Instead, melanic patterns expanded as expected and scale structures were altered across all color of scales (Figure 3). The melanic scales of Heparin treated female wings had an ultrastructure that was distinct from both black and yellow scales of control wings, with triple pronged leading edges that are not commonly found across Z. cesonia wings (Figure 3, blue box).
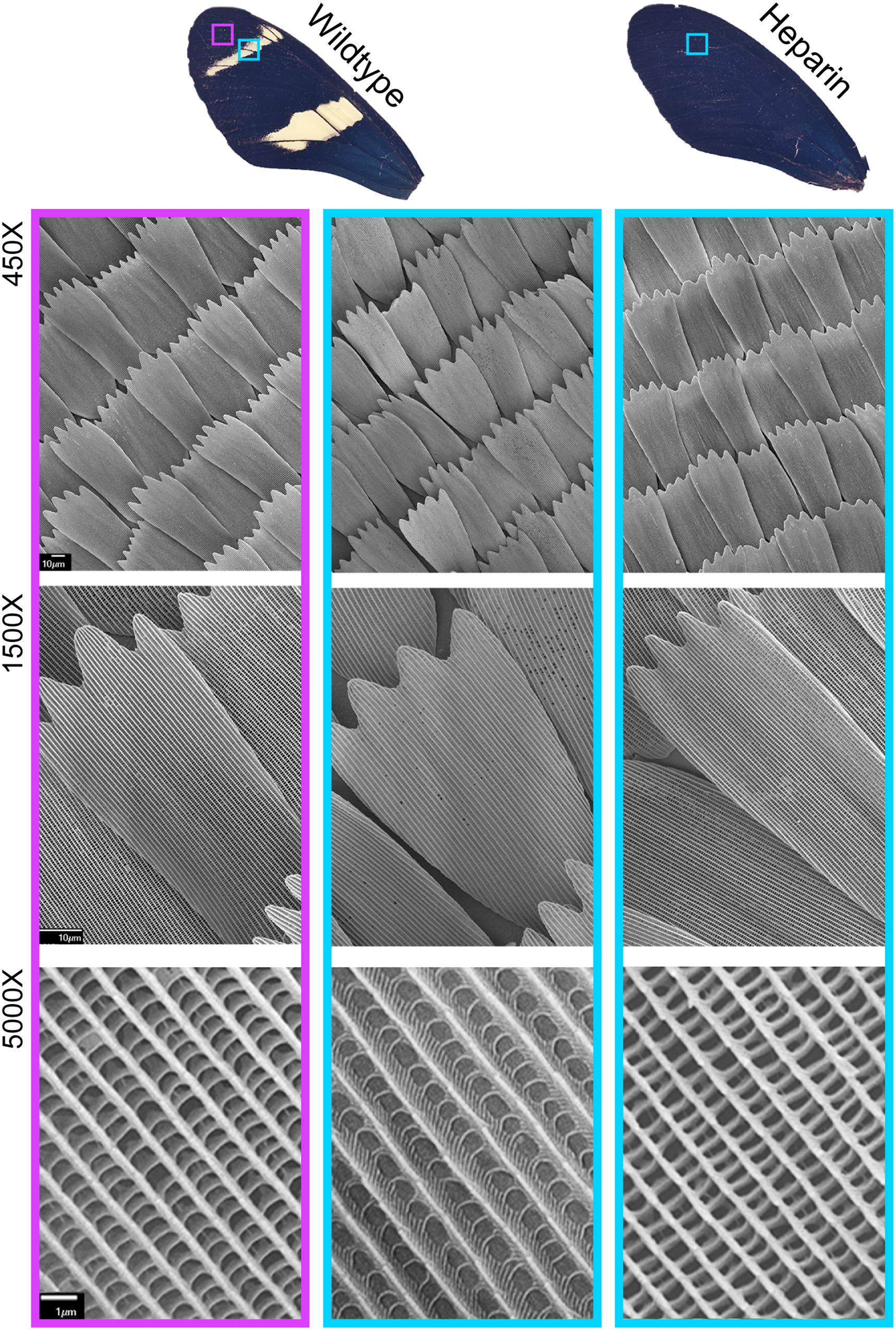
Figure 4. Heparin impacts both scale pigmentation and structure in H. sara. Wings from Martin et al. (2012). Heparin injections result in yellow-to-blue/black scale color shifts, and SEM imaging reveals concordant changes in ultrastructure (5000X).
It is worth noting the large difference in Heparin dosage among nymphalid and pierid butterfly species; for example, 5 μg of Heparin was required to observe wing pattern changes in Heliconius, while only 0.5 μg was used in Z. cesonia, and higher dosage always resulted in pupal mortality (n = 25). This suggests that pierids may have a more sensitive response to Heparin, possibly due to Heparin binding to different ligands in Z. cesonia than in H. sara. Collectively, these Heparin results show that although Heparin appeared to produced similar wing pattern phenotypes across butterfly families, the effects of Heparin on scale development and dosage responses differ dramatically. Collectively, these findings suggest that targets of Heparin impact both scale pigmentation and structure across nymphalid and pierid butterfly wings, but also indicate that pierids may use a different set of Wnts or other signaling molecules to pre-pattern wing coloration.
Discussion
Wnts in Butterfly Wing Pattern: Conservation and Divergence
Wnt gene content, clustering, and amino acid sequence appear to be highly conserved among pierids, as well as other Lepidoptera. However, the deployment of these genes appears to be highly divergent among different butterfly lineages. Interestingly several Wnt genes displayed divergent expressions between developing wings of nymphalid and pierid butterflies. The expression profiles for Wnt genes in Z. cesonia clearly showed that they do not deploy the same Wnt genes as nymphalids in wing pattern development.
Most surprising was the lack of WntA expression during wing development in Z. cesonia, despite its importance in wing pattern development of nymphalid butterflies (Mazo-Vargas et al., 2017; Concha et al., 2019). In nymphalids, WntA expression in larval wing discs prefigures adult wing elements and diminishes during the first days AFP (Martin et al., 2012; Martin and Reed, 2014). Z. cesonia completely lacked WntA expression, suggesting that this pierid lacks the WntA-positive stripe system of nymphalids called the Central Symmetry System (CSS). Of note, classic work in comparative morphology has established that this pattern is divergent and unusually reduced in Pieridae (Schwanwitsch, 1956) and it will be interesting to further investigate if WntA expression has been completely lost in additional pierids to test for generalization. It is possible that WntA may be expressed in levels too low to detect in the Z. cesonia transcriptome, with an average of 18.9 million reads per library, but if so such low levels of expression, it is unlikely that WntA would be serving the same vital function in Z. cesonia as it is in nymphalid butterflies where it is more highly expressed.
Differences in Wnt gene deployment during wing development among lepidoptera lineages likely result from changes in regulatory regions of the Wnt genes. Across Lepidoptera, there is evidence of strong purifying selection on Wnt gene amino acid sequences, demonstrating that both Wnt gene content and sequences are highly conserved (Martin et al., 2012; Mazo-Vargas et al., 2017; Holzem et al., 2019). In Heliconius butterflies, differences in the spatial expression of WntA across developing wings are associated with genetic changes in non-coding regions near WntA. Van Belleghem et al. (2017) speculate that these reflect putative cis-regulatory elements (CREs) that are responsible for initiating WntA expression in narrow wing regions that correspond to pattern elements in the adult wings. Similarly, Van Belleghem et al. identified CREs near the gene optix that were associated with red color patterning in Heliconius, which (Lewis et al., 2019) recently used CRISPR to edit the CREs and demonstrate their function in red pattern development. We suggest that evolution of CREs near the Wnt gene clusters is likely responsible for differences in Wnt gene deployment and pattern divergence of lepidoptera wings.
Wnt Pathway Involved in Structural Coloration
Heparin treatments consistently lead to an expansion of melanism across butterfly wings. However, if Heparin is impacting melanism via the Wnt pathway, it is likely affecting different Wnt ligands in different butterfly species. In nymphalid butterflies, WntA has been shown to be spatially expressed in wing regions that melanism expands and is a likely candidate target of Heparin treatments. However, in Z. cesonia wing discs, WntA is not expressed, and at the time of Heparin injections only Wnt1, Wnt6, and possibly Wnt7 ligands appear to be present in wing tissue. Also different from nymphalid butterflies, the expansion of melanism was greatest in the anterior portion of the wing in Z. cesonia and C. eurytheme, where in nymphalids, melanic expansion largely occurred in elements of the CSS of the wing (Martin et al., 2012). Lastly, we note that the melanic response to Heparin was sex dependent, with females exhibiting greater expansions than males. This sexual dimorphism has not been observed in nymphalids; however, in species such as Heliconius, sexual dimorphism in wing color pattern is rare. Among pierids, wing color patterns are often sexually dimorphic, most notably in UV reflective patterns which are often present in males, but not females, as is found in Z. cesonia (Silberglied and Taylor, 1973; Stella et al., 2018; Fenner et al., 2019). Interestingly, in Drosophila Wnt7 expression is sexually dimorphic and regulated by doublesex during embryonic gonad development of male specific pigment cells (Deshpande et al., 2016). Collectively, the gene expression and Heparin treatment results suggest that Wnt7 could be a candidate ligand involved in sexually dimorphic wing development in a pierid butterfly.
Previous studies using Heparin assays have largely focused on pattern level changes across the wing, here we show how Heparin impacts the development of the wing scales. The Heparin assays suggest that the Wnt pathway may play a role in the development of structural as well as pigment coloration. Although much is known about the genes responsible for pigment colors such as melanin, little is known about the pathways responsible for generating structural colors (Zhang et al., 2017). Here we show that males lose the ability to produce their UV patterns, but not due to cell fate changes, but do to overall scale shape changes. In nymphalid butterflies, WntA has recently been implemented as a modulator of both scale structure and color (Concha et al., 2019). Since WntA is not expressed in Z. cesonia, but both scale and pattern phenotypes were altered, this suggests that the Wnt pathway in general may play an important role in the development of both scale coloration and overall structure.
Multiple Pathways Involved in Wing Pattern Development
To date, there are at least three different Wnt signaling pathways: the canonical Wnt/beta pathway, the non-canonical Wnt/JNK (Wnt/PCP), and the Wnt/Ca2+ pathway (Nusse, 2012). In the last decade, it has become evident that two or more of these Wnt signaling pathways often signal within the same cell or territories to influence cell fate (Range et al., 2013; Range, 2018; Martínez-Bartolomé and Range, 2019). Here we characterized the expression of four Wnt genes in Z. cesonia throughout pupal wing development. As stated above, our Heparin injections showed perturbations in both scale color, which is likely due to transcriptional regulation, and scale structure, which is due to subcellular changes in the actin-myosin cytoskeleton. In many cases, the non-canonical Wnt signaling pathways are involved in morphogenetic processes; therefore, it is possible that multiple Wnt pathways are activated during wing development. We realize that individual Wnts can activate canonical and/or non-canonical pathways depending on the cellular context; it is interesting to note that Wnt11 has been shown to activate the Wnt/JNK pathway in Drosophila development (Uysal-Onganer and Kypta, 2012). Although the expression of Wnt11 is low during wing development in Z. cesonia, its peak expression occurs during days 2 and 3 when scale formation is taking place. On the other hand, Wnt7 expression during wing development, like WntA, has only been seen in butterfly wings and thus the Wnt pathways they may initiate remain unresolved. Together, the handful of Wnt genes expressed suggest that multiple Wnt pathways could be concordantly regulating various aspects of wing development. Further characterization needs to be conducted in order to address which Wnt pathways are deployed during wing development in Z. cesonia.
In addition, other signaling pathways are likely involved and may contribute to the heparin-induced wing changes. Heparin binding is not limited to the Wnt family and impact the diffusion of other signaling genes, such as fibroblast growth factor (FGF), Hedgehog (Hh), and transforming growth factor-β (TGFβ) families. In Z. cesonia Hh, TGF-β activated kinase 1 (Tak1), branchless (FGF family), and decapentaplegic (dpp) (FGF family) are all expressed during wing development and could have been impacted by heparin treatments. In Junonia butterflies, Hh has been implicated in eyespot pattern development (Tong et al., 2012); however, the morphogens Tak1, branchless, and dpp have yet to be investigated in another butterfly species. Therefore, although Wnt genes have played a major role in Nymphalid wing pattern development and diversification (Concha et al., 2019), the results of this paper suggest that Pierids express Wnts differently and further characterization is needed for other families of morphogens that may be involved in wing patterning.
Data Availability Statement
The names of the repository/repositories and accession number(s) can be found at: NCBI [BIOPROJECT PRJNA587792], GitHub [https://github.com/bcount/Colias10X].
Author Contributions
JF and BC designed the study, conducted the pharmaceutical assays, and wrote the manuscript draft. JF collected microscopy and wing image data and prepared RNA-seq libraries. CB and LR-C conducted RNA-seq analyses. RP provided sequencing of the RNA-seq libraries. AR conducted pharmaceutical assays for C. eurytheme. FH conducted phylogenetic and molecular evolution analyses. AM and RR contributed to experimental design and data interpretation. All authors contributed to preparation and editing of the manuscript.
Funding
This research was funded by the National Science Foundation awards IOS-1755329 to AM and BC, OIA-1736026 to RP, FH, RR, and BC, IOS-1656389 to RP and AM, and Texas Ecolabs awards to BC.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jared Cole for assistance with RNA-seq library preparation, and Danilo Gusmao Ferreira Souto for assistance with Heparin injections and butterfly rearing. We thank the MSU I2AT for technical assistance with SEM. We also thank the University of Puerto Rico, Rio Piedras Sequencing and Genomics Facility, and the UPR High Performance Computing Facility for assistance with the generation and analyses of RNA-seq data.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.00197/full#supplementary-material
Footnotes
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/s0022-2836(05)80360-2
Anisimova, M., Gil, M., Dufayard, J.-F., Dessimoz, C., and Gascuel, O. (2011). Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 60, 685–699. doi: 10.1093/sysbio/syr041
Baker, N. E. (1987). Molecular cloning of sequences from wingless, a segment polarity gene in Drosophila: the spatial distribution of a transcript in embryos. EMBO J. 6, 1765–1773. doi: 10.1002/j.1460-2075.1987.tb02429.x
Baker, N. E. (1988). Transcription of the segment-polarity gene wingless in the imaginal discs of Drosophila, and the phenotype of a pupal-lethal wg mutation. Development 102, 489–497.
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bolognesi, R., Beermann, A., Farzana, L., Wittkopp, N., Lutz, R., Balavoine, G., et al. (2008). Tribolium Wnts: evidence for a larger repertoire in insects with overlapping expression patterns that suggest multiple redundant functions in embryogenesis. Dev. Genes Evol. 218, 193–202. doi: 10.1007/s00427-007-0170-3
Challi, R. J., Kumar, S., Dasmahapatra, K. K., Jiggins, C. D., and Blaxter, M. (2016). Lepbase: the Lepidopteran genome database. bioRxiv [Preprint]. doi: 10.1101/056994
Chen, S., Huang, T., Zhou, Y., Han, Y., Xu, M., and Gu, J. (2017). AfterQC: automatic filtering, trimming, error removing and quality control for fastq data. BMC Bioinformatics 18:80. doi: 10.1186/s12859-017-1469-3
Chen, Z., Zhu, J. Y., Fu, Y., Richman, A., and Han, Z. (2016). Wnt4 is required for ostia development in the Drosophila heart. Dev Biol. 413, 188–198. doi: 10.1016/j.ydbio.2016.03.008
Cohen, E. D., Mariol, M.-C., Wallace, R. M. H., Weyers, J., Kamberov, Y. G., Pradel, J., et al. (2002). DWnt4 regulates cell movement and focal adhesion kinase during Drosophila ovarian morphogenesis. Dev. Cell 2, 437–448. doi: 10.1016/s1534-5807(02)00142-9
Concha, C., Wallbank, R. W. R., Hanly, J. J., Fenner, J., Livraghi, L., Rivera, E. S., et al. (2019). Interplay between developmental flexibility and determinism in the evolution of mimetic heliconius wing patterns. Curr. Biol. 23, 3996.e4–4009.e4. doi: 10.1016/j.cub.2019.10.010
Deshpande, G., Nouri, A., and Schedl, P. (2016). Wnt signaling in sexual dimorphism. Genetics 202, 661–673. doi: 10.1534/genetics.115.177857
Ding, X., Liu, J., Zheng, L., Song, J., Li, N., Hu, H., et al. (2019). Genome-wide identification and expression profiling of wnt family genes in the silkworm, Bombyx mori. Int. J. Mol. Sci. 20:1221. doi: 10.3390/ijms20051221
Edayoshi, M., Yamaguchi, J., and Fujiwara, H. (2015). Protruding structures on caterpillars are controlled by ectopic Wnt1 expression. PLoS One 10:e0121736. doi: 10.1371/journal.pone.0121736
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Fenner, J., Rodriguez-Caro, L., and Counterman, B. (2019). Plasticity and divergence in ultraviolet reflecting structures on Dogface butterfly wings. Arthropod. Struct. Dev. 51, 14–22. doi: 10.1016/j.asd.2019.06.001
Gallant, J. R., Imhoff, V. E., Martin, A., Savage, W. K., Chamberlain, N. L., Pote, B. L., et al. (2014). Ancient homology underlies adaptive mimetic diversity across butterflies. Nat. Commun. 5, 1–10.
Goldman, N., and Yang, Z. (1994). A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11, 725–736.
Hanly, J. (2017). Developmental Basis of Wing Pattern Diversity in Heliconius Butterflies. Doctoral thesis, Cambridge University, Cambridge.
Hanly, J. J., Wallbank, R. W. R., Owen McMillan, W., and Jiggins, C. D. (2019). Conservation and flexibility in the gene regulatory landscape of heliconiine butterfly wings. EvoDevo 10:15. doi: 10.1186/s13227-019-0127-4
Hoang, D. T., Vinh, L. S., Flouri, T., Stamatakis, A., von Haeseler, A., and Minh, B. Q. (2018). MPBoot: fast phylogenetic maximum parsimony tree inference and bootstrap approximation. BMC Evol. Biol. 18:11. doi: 10.1186/s12862-018-1131-3
Holzem, M., Braak, N., Brattström, O., McGregor, A. P., and Breuker, C. J. (2019). Wnt gene expression during early embryogenesis in the nymphalid butterfly Bicyclus anynana. Front. Ecol. Evol. 7:468. doi: 10.3389/fevo.2019.00468
Howe, K. L., Contreras-Moreira, B., De Silva, N., Maslen, G., Akanni, W., Allen, J., et al. (2020). Ensembl Genomes 2020-enabling non-vertebrate genomic research. Nucleic Acids Res. 48, D689–D695.
Janson, K., Cohen, E. D., and Wilder, E. L. (2001). Expression of DWnt6, DWnt10, and DFz4 during Drosophila development. Mech. Dev. 103, 117–120. doi: 10.1016/s0925-4773(01)00323-9
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kawahara, A. Y., Plotkin, D., Espeland, M., Meusemann, K., Toussaint, E. F. A., Donath, A., et al. (2019). Phylogenomics reveals the evolutionary timing and pattern of butterflies and moths. Proc. Natl. Acad. Sci. U.S.A. 116, 22657–22663. doi: 10.1073/pnas.1907847116
Koshikawa, S., Giorgianni, M. W., Vaccaro, K., Kassner, V. A., Yoder, J. H., Werner, T., et al. (2015). Gain of cis-regulatory activities underlies novel domains of wingless gene expression in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 112, 7524–7529. doi: 10.1073/pnas.1509022112
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kurtz, S., Phillippy, A., Delcher, A. L., Smoot, M., Shumway, M., Antonescu, C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5:R12.
Lewis, J. J., Geltman, R. C., Pollak, P. C., Rondem, K. E., Van Belleghem, S. M., Hubisz, M. J., et al. (2019). Parallel evolution of ancient, pleiotropic enhancers underlies butterfly wing pattern mimicry. Proc. Natl. Acad. Sci. U.S.A. 116, 24174–24183. doi: 10.1073/pnas.1907068116
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Macdonald, W. P., Martin, A., and Reed, R. D. (2010). Butterfly wings shaped by a molecular cookie cutter: evolutionary radiation of lepidopteran wing shapes associated with a derived Cut/wingless wing margin boundary system. Evol. Dev. 12, 296–304. doi: 10.1111/j.1525-142x.2010.00415.x
Martin, A., Papa, R., Nadeau, N. J., Hill, R. I., Counterman, B. A., Halder, G., et al. (2012). Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand. Proc. Natl. Acad. Sci. U.S.A. 109, 12632–12637. doi: 10.1073/pnas.1204800109
Martin, A., and Reed, R. D. (2010). Wingless and aristaless2 define a developmental ground plan for moth and butterfly wing pattern evolution. Mol. Biol. Evol. 27, 2864–2878. doi: 10.1093/molbev/msq173
Martin, A., and Reed, R. D. (2014). Wnt signaling underlies evolution and development of the butterfly wing pattern symmetry systems. Dev. Biol. 395, 367–378. doi: 10.1016/j.ydbio.2014.08.031
Martínez-Bartolomé, M., and Range, R. C. (2019). A biphasic role of non-canonical Wnt16 signaling during early anterior-posterior patterning and morphogenesis of the sea urchin embryo. Development 146:dev168799. doi: 10.1242/dev.168799
Mazo-Vargas, A., Concha, C., Livraghi, L., Massardo, D., Wallbank, R. W. R., Zhang, L., et al. (2017). Macroevolutionary shifts of WntA function potentiate butterfly wing-pattern diversity. Proc. Natl. Acad. Sci. U.S.A. 114, 10701–10706. doi: 10.1073/pnas.1708149114
Murat, S., Hopfen, C., and McGregor, A. P. (2010). The function and evolution of Wnt genes in arthropods. Arthropod. Struct. Dev. 39, 446–452. doi: 10.1016/j.asd.2010.05.007
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nusse, R. (2012). Wnt signaling. Cold Spring Harb. Perspect. Biol. 4:a011163. doi: 10.1101/cshperspect.a011163
Papke, R., Darrell, S., Kemp, J., and Rutowski, R. L. (2007). Multimodal signalling: structural ultraviolet reflectance predicts male mating success better than pheromones in the butterfly Colias eurytheme L.(Pieridae). Anim. Behav. 73, 47–54. doi: 10.1016/j.anbehav.2006.07.004
Picelli, S., Björklund, A. K., Reinius, B., Sagasser, S., Winberg, G., and Sandberg, R. (2014). Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 24, 2033–2040. doi: 10.1101/gr.177881.114
Range, R. C. (2018). Canonical and non-canonical Wnt signaling pathways define the expression domains of Frizzled 5/8 and Frizzled 1/2/7 along the early anterior-posterior axis in sea urchin embryos. Dev. Biol. 444, 83–92. doi: 10.1016/j.ydbio.2018.10.003
Range, R. C., Angerer, R. C., and Angerer, L. M. (2013). Integration of canonical and noncanonical Wnt signaling pathways patterns the neuroectoderm along the anterior–posterior axis of sea urchin embryos. PLoS Biol. 11:e1001467. doi: 10.1371/journal.pbio.1001467
Rao, T. P., and Kühl, M. (2010). An updated overview on Wnt signaling pathways: a prelude for more. Circ. Res. 106, 1798–1806. doi: 10.1161/circresaha.110.219840
Rodriguez-Caro, L., Fenner, J., Benson, C., Van Belleghem, S. M., and Counterman, B. A. (2020). Genome assembly of the dogface butterfly Zerene cesonia. Genome Biol. Evol. 12, 3580–3585. doi: 10.1093/gbe/evz254
Schwanwitsch, B. N. (1956). Wing pattern of pierid butterflies (Pieridae, Lepidoptera), Ent Obozr, Moscow. Entomol. Obozrenie 35, 285–301.
Serfas, M. S., and Carroll, S. B. (2005). Pharmacologic approaches to butterfly wing patterning: sulfated polysaccharides mimic or antagonize cold shock and alter the interpretation of gradients of positional information. Dev. Biol. 287, 416–424. doi: 10.1016/j.ydbio.2005.09.014
Shelby, E. A., Fenner, J., and Counterman, B. A. (2020). Impact of artificial diet on development and coloration in Zerene cesonia. J. Lepid. Soc. 74, 65–72. doi: 10.18473/lepi.74i2.a1
Silberglied, R. E., and Taylor, O. R. (1973). Ultraviolet differences between the sulphur butterflies, Colias eurytheme and C. philodice, and a possible isolating mechanism. Nature 241, 406–408. doi: 10.1038/241406a0
Somorjai, I. M. L., Martí-Solans, J., Diaz-Gracia, M., Nishida, H., Imai, K. S., Escrivà, H., et al. (2018). Wnt evolution and function shuffling in liberal and conservative chordate genomes. Genome Biol. 19:98.
Sourakov, A. (2018). Scientific Note: The Emperor’s new clothes: radical transformation of the wing pattern in Asterocampa clyton caused by heparin. Trop. Lepid. Res. 28, 29–31.
Stella, D., Faltýnek Fric, Z., Rindoš, M., Kleisner, K., and Pechácek, P. (2018). Distribution of ultraviolet ornaments in colias butterflies (Lepidoptera: Pieridae). Environ. Entomol. 47, 1344–1354. doi: 10.1093/ee/nvy111
Tomoyasu, Y., Arakane, Y., Kramer, K. J., and Denell, R. E. (2009). Repeated co-options of exoskeleton formation during wing-to-elytron evolution in beetles. Curr. Biol. 19, 2057–2065. doi: 10.1016/j.cub.2009.11.014
Tong, X., Lindemann, A., and Monteiro, A. (2012). Differential involvement of Hedgehog signaling in butterfly wing and eyespot development. PLoS One 7:e51087. doi: 10.1371/journal.pone.0051087
Trifinopoulos, J., Nguyen, L.-T., von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Uysal-Onganer, P., and Kypta, R. M. (2012). Wnt11 in 2011–the regulation and function of a non-canonical Wnt. Acta Physiol. 204, 52–64. doi: 10.1111/j.1748-1716.2011.02297.x
Van Belleghem, S. M., Rastas, P., Papanicolaou, A., Martin, S. H., Arias, C. F., Supple, M. A., et al. (2017). Complex modular architecture around a simple toolkit of wing pattern genes. Nat. Ecol. Evol. 1:52.
Weisenfeld, N. I., Kumar, V., Shah, P., Church, D. M., and Jaffe, D. B. (2017). Direct determination of diploid genome sequences. Genome Res. 27, 757–767. doi: 10.1101/gr.214874.116
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Zhang, L., Martin, A., Perry, M. W., van der Burg, K. R. L., Matsuoka, Y., Monteiro, A., et al. (2017). Genetic basis of melanin pigmentation in butterfly wings. Genetics 205, 1537–1550. doi: 10.1534/genetics.116.196451
Keywords: pigmentation, ultraviolet, Lepidoptera, RNA-seq, wing development
Citation: Fenner J, Benson C, Rodriguez-Caro L, Ren A, Papa R, Martin A, Hoffmann F, Range R and Counterman BA (2020) Wnt Genes in Wing Pattern Development of Coliadinae Butterflies. Front. Ecol. Evol. 8:197. doi: 10.3389/fevo.2020.00197
Received: 09 March 2020; Accepted: 29 May 2020;
Published: 30 June 2020.
Edited by:
Marcus Kronforst, The University of Chicago, United StatesReviewed by:
Sean Mullen, Boston University, United StatesLuana S. Maroja, Williams College, United States
Copyright © 2020 Fenner, Benson, Rodriguez-Caro, Ren, Papa, Martin, Hoffmann, Range and Counterman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brian A. Counterman, bcounterman@biology.msstate.edu
 Jennifer Fenner
Jennifer Fenner Caleb Benson2
Caleb Benson2  Luis Rodriguez-Caro
Luis Rodriguez-Caro Anna Ren
Anna Ren Federico Hoffmann
Federico Hoffmann