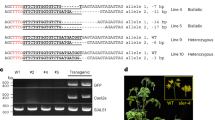
Abstract
Conditional manipulation of gene expression is a key approach to investigating the primary function of a gene in a biological process. While conditional and cell-type-specific overexpression systems exist for plants, there are currently no systems available to disable a gene completely and conditionally. Here, we present a new tool with which target genes can efficiently and conditionally be knocked out by genome editing at any developmental stage. Target genes can also be knocked out in a cell-type-specific manner. Our tool is easy to construct and will be particularly useful for studying genes having null alleles that are non-viable or show pleiotropic developmental defects.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
Vectors created in this study have been deposited in Addgene for distribution. Addgene ID numbers are presented in Fig. 1 and Extended Data Fig. 1. All plant material, expression constructs and data supporting findings of this study are available from the corresponding author upon request. Source data are provided with this paper.
References
Candela, H., Perez-Perez, J. M. & Micol, J. L. Uncovering the post-embryonic functions of gametophytic- and embryonic-lethal genes. Trends Plant Sci. 16, 336–345 (2011).
Borghi, L. et al. Arabidopsis RETINOBLASTOMA-RELATED is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell 22, 1792–1811 (2010).
Guo, J., Wei, J., Xu, J. & Sun, M. X. Inducible knock-down of GNOM during root formation reveals tissue-specific response to auxin transport and its modulation of local auxin biosynthesis. J. Exp. Bot. 65, 1165–1179 (2014).
Liu, L. & Chen, X. Intercellular and systemic trafficking of RNAs in plants. Nat. Plants 4, 869–878 (2018).
Heidstra, R., Welch, D. & Scheres, B. Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 18, 1964–1969 (2004).
Wachsman, G., Heidstra, R. & Scheres, B. Distinct cell-autonomous functions of RETINOBLASTOMA-RELATED in Arabidopsis stem cells revealed by the Brother of Brainbow clonal analysis system. Plant Cell 23, 2581–2591 (2011).
Wildwater, M. et al. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123, 1337–1349 (2005).
Qi, Y. P. et al. Targeted deletion and inversion of tandemly arrayed genes in Arabidopsis thaliana using zinc finger nucleases. G3 3, 1707–1715 (2013).
Christian, M., Qi, Y. P., Zhang, Y. & Voytas, D. F. Targeted mutagenesis of Arabidopsis thaliana using engineered TAL effector nucleases. G3 3, 1697–1705 (2013).
Mao, Y. F., Botella, J. R., Liu, Y. G. & Zhu, J. K. Gene editing in plants: progress and challenges. Natl Sci. Rev. 6, 421–437 (2019).
Decaestecker, W. et al. CRISPR-TSKO: a technique for efficient mutagenesis in specific cell types, tissues, or organs in Arabidopsis. Plant Cell 31, 2868–2887 (2019).
Ma, X. et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284 (2015).
Siligato, R. et al. MultiSite gateway-compatible cell type-specific gene-inducible system for plants. Plant Physiol. 170, 627–641 (2016).
Zuo, J., Niu, Q. W. & Chua, N. H. Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24, 265–273 (2000).
Karimi, M., Inze, D. & Depicker, A. GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 (2002).
Aida, M. et al. The PHETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119, 109–120 (2004).
Galinha, C. et al. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449, 1053–1057 (2007).
Mähönen, A. P. et al. PLETHORA gradient formation mechanism separates auxin responses. Nature 515, 125–129 (2014).
Vandenberg, C., Willemsen, V., Hage, W., Weisbeek, P. & Scheres, B. Cell fate in the Arabidopsis root meristem determined by directional signaling. Nature 378, 62–65 (1995).
Brinkman, E. K., Chen, T., Amendola, M. & van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014).
Ebel, C., Mariconti, L. & Gruissem, W. Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429, 776–780 (2004).
Cruz-Ramirez, A. et al. A SCARECROW-RETINOBLASTOMA protein network controls protective quiescence in the Arabidopsis root stem cell organizer. PLoS Biol. 11, e1001724 (2013).
Geldner, N. et al. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112, 219–230 (2003).
Kleine-Vehn, J. et al. ARF-GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Curr. Biol. 18, 526–531 (2008).
Steinmann, T. et al. Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286, 316–318 (1999).
Geldner, N. et al. Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development 131, 389–400 (2004).
Shevell, D. E. et al. Emb30 is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7. Cell 77, 1051–1062 (1994).
Scarpella, E., Marcos, D., Friml, J. & Berleth, T. Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20, 1015–1027 (2006).
Fulcher, N. & Sablowski, R. Hypersensitivity to DNA damage in plant stem cell niches. Proc. Natl Acad. Sci. USA 106, 20984–20988 (2009).
Horvath, B. M. et al. Arabidopsis RETINOBLASTOMA-RELATED directly regulates DNA damage responses through functions beyond cell cycle control. EMBO J. 36, 1261–1278 (2017).
Carbonell, A. et al. New generation of artificial MicroRNA and synthetic trans-acting small interfering RNA vectors for efficient gene silencing in Arabidopsis. Plant Physiol. 165, 15–29 (2014).
Schwab, R., Ossowski, S., Riester, M., Warthmann, N. & Weigel, D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18, 1121–1133 (2006).
Matos, J. L. et al. Irreversible fate commitment in the Arabidopsis stomatal lineage requires a FAMA and RETINOBLASTOMA-RELATED module. eLife 3, e03271 (2014).
Brand, L. et al. A versatile and reliable two-component system for tissue-specific gene induction in Arabidopsis. Plant Physiol. 141, 1194–1204 (2006).
Moore, I., Samalova, M. & Kurup, S. Transactivated and chemically inducible gene expression in plants. Plant J. 45, 651–683 (2006).
Kareem, A. et al. Protocol: a method to study the direct reprogramming of lateral root primordia to fertile shoots. Plant Methods 12, 27 (2016).
Ursache, R., Andersen, T. G., Marhavy, P. & Geldner, N. A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. Plant J. 93, 399–412 (2018).
Bargmann, B. O. & Birnbaum, K. D. Fluorescence activated cell sorting of plant protoplasts. J. Vis. Exp. 36, e1673 (2010).
Edwards, K., Johnstone, C. & Thompson, C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19, 1349 (1991).
Smetana, O. et al. High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature 565, 485–489 (2019).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at http://arxiv.org/abs/1303.3997 (2013).
McKenna, A. et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Poplin, R. et al. Scaling accurate genetic variant discovery to tens of thousands of samples. Preprint at https://doi.org/10.1101/201178 (2017).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Acknowledgements
We thank B. Scheres (Wageningen University) and N. Geldner (University of Lausanne) for sharing published materials; N. Geldner for providing support for R.U.; K. D. Birnbaum (University of New York) for helpful discussions; T. Pessa-Morikawa (University of Helsinki) for technical support on FACS; A. Vaten (University of Helsinki) for cotyledon epidermis imaging; and S. el-Showk for proofreading of the manuscript. This work was supported by the Academy of Finland (grant nos. 316544, 266431, 307335), the European Research Council (ERC-CoG CORKtheCAMBIA, agreement no. 819422), the University of Helsinki HiLIFE fellowship (to X.W., L.Y., M.L. and A.P.M.) and the European Molecular Biology Organisation (EMBO ALTF, no. 1046-2015 to R.U.). X.W. is also supported by a grant from the Chinese Scholarship Council.
Author information
Authors and Affiliations
Contributions
X.W. and A.P.M. designed the experiments. X.W. conducted all experiments, but L.Y. carried out the analysis for Supplementary Table 1 and M.L. performed FACS. R.U. generated and tested the new destination vectors. A.L. determined the indel mutation efficiency of amplicon deep sequencing. X.W. and A.P.M. analysed the results and wrote the manuscript, with input from all co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Yiping Qi, Kejian Wang, Huawei Zhang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Non-destructive screening markers facilitate identification of transformed seeds.
a, Non-destructive fluorescent screening destination vectors generated in this study. b, Examples of transgenic seeds containing pFRm43GW screened under the fluorescence-binocular in the T1 (left) and T2 (right) generations. Experiments in (b) have been repeated more than three times.
Extended Data Fig. 2 IGE system enables real time observation of genome editing.
To monitor PLT2 editing dynamics, a time-course 17-β induction was conducted to ipWER»Cas9p-tagRFP-PLT2 in gPLT2-3xYFP; plt1,2 (T2 generation, line #1). Cas9p-tagRFP fluorescence appeared after 4 hours of induction, followed by gradual reduction of PLT2-3xYFP expression (starting after 12 hours of induction). Cas9p-tagRFP expression and editing activity was gradually spread inwards, likely due to the radial diffusion of 17-β within ipWER domain. White dotted lines mark the RM outlines. Cell walls are visualized by calcofluor. Experiments were repeated three times. Numbers indicate the frequency of observed phenotype within given induction duration. Scale bar, 50 μm.
Extended Data Fig. 3 Detection and quantification of PLT2 deletion from sorted cell populations.
a, PCR-based detection of PLT2 deletion in sorted cell populations. While several truncated bands were visible, the predominant truncated band corresponds to the large fragment deletion between target1 and target4 (see location of target sites in Supplementary Fig. 1). Experiments were repeated three times. b, qPCR primer design strategy for PLT2 deletion efficiency quantification. To avoid amplification of native PLT2, forward primer (F) was designed at attB1 site linking promoter and genomic PLT2 and reverse primer (R) was designed at downstream of target1. c, Quantification of PLT2 deletion efficiency by qPCR with the pooled genome DNA from the sorted population (n > 600) as template. Error bars represent s.d., and experiments were repeated three times with similar results. Individual values (black dots) and means (bars) are shown. Ctrl indicates the gPLT2-3xYFP; plt1,2.
Extended Data Fig. 4 sgRNA promoter identity affects editing efficiency in Arabidopsis roots.
For each construct, the indicated sgRNA promoter was used to drive transcription of sgRNA1, while ip35S was used to guide Cas9p transcription. AtU3b and AtU6-29 showed the highest editing efficiency in T1 seedlings after one-day of induction (1d 17-β). This may explain the preferred detection of deletion between target1 (AtU3b) and target4 (AtU6-29) when four sgRNAs were used in a single construct (Supplementary Fig. 4 and Extended Data Fig. 3a). Transcription of transfer RNA, together with sgRNA1 under the AtU3b promoter, also resulted in efficient PLT2 editing. White dotted lines mark the region with reduced YFP signal. This corresponds to the region where ip35S is active (Fig. 2a and Supplementary Fig. 3). Cell walls are highlighted by calcofluor. Numbers indicate the frequency of similar results in the independent T1 samples analysed. All experiments were repeated three times. Scale bar, 50 μm.
Extended Data Fig. 5 RBR functions cell-autonomously in the RM.
a, A three-day mock treatment of ip35S»Cas9p-RBR in RBR-YFP; amiGORBR. b, A one-day induction caused a reduced RBR-YFP signal mainly in the root cap region without an obvious phenotype. c, A three-day induction of RBR editing with ip35S typically led to LRC overproliferation (white arrows) without affecting the YFP signal in other domains. While half of the transformants showed sectors of variable size lacking RBR-YFP expression (left panel in c), the other half showed almost complete absence of RBR-YFP in the domain of ip35S (right panel). Cell walls are visualized by calcofluor. Numbers indicate the frequency of the observed phenotype in independent T1 samples. Experiments were repeated three times. Scale bar, 50 μm.
Extended Data Fig. 6 Post-embryonically inducing GNOM editing recapitulates the phenotypes of the gnom mutant.
a, Plants with ipWOL»Cas9p-tagRFP-GNOM in PIN1-GFP after ten days germination on mock or 17-β plates. Inducing GNOM editing led to shorter roots, agravitropic growth and decreased lateral root (LR) numbers. Adventitious roots from the hypocotyl were frequently found, however these roots were not counted in LR quantification. For each independent root, LR number and root length is plotted in (b). Experiments were repeated three times. Scale bar, 1 cm.
Extended Data Fig. 7 GNOM is required for PIN1 polarity and expression.
a, GNOM expression disappeared from the vasculature after a 6-day induction of ipWOL»Cas9p-tagRFP-GNOM in GN-GFP. Due to the weak GFP signal, only roots showing a clear loss of GFP signal were included in quantification. b, A three-day induction of ipWOL»Cas9p-tagRFP-GNOM in PIN1-GFP resulted in loss of polarity and decreased expression of PIN1-GFP in the endodermis (en), pericycle (p) and stele (s) (white arrows). Right panels are magnified images of the regions marked with a red box in the left panels. Cell walls are marked by calcofluor. Numbers indicate the frequency of the observed phenotype in independent T1 samples analyzed. Experiments were repeated three times. Scale bar in right panels of a, 25 μm; others, 50 μm.
Extended Data Fig. 8 Cas9p-mediated genome editing in proximal stem cells induces cell death.
a, Stem cell death surrounding the QC was observed after one-day induction of ip35S»Cas9p-PLT2. Based on cell types, the cell death response is classified into three categories: provascular cell death, LRC/epidermis initial cell death and columella initial cell death. Samples were counted twice if they had cell death in two different categories. b, Cell death of provascular cells and early descendants was induced after one-day induction of ipWOL»Cas9p-tagRFP-PLT2/RBR/GNOM. Cell walls are highlighted by propidium iodide (PI). Under PI detection settings, Cas9p-tagRFP is also visible. Numbers indicate the frequency of the observed phenotype in independent T1 samples analyzed. Experiments were repeated three times. Scale bars, 50 μm.
Extended Data Fig. 9 A single IGE construct targeting a gene encoding a fluorescent reporter has the potential to disrupt different transgene targets.
a, Editing YFP instead of PLT2 in the ipWER expression region caused changes similar to direct PLT2 editing. The RM had fewer LRC layers (white arrowheads), as well as premature expansion of epidermal cells and a broad, faint YFP signal. The Cas9p-tagRFP signal is frequently invisible. b, Editing YFP led to QC (black arrow) differentiation at a lower frequency. (c) Targeting the YFP of RBR-YFP in the LRC led to LRC overproliferation, similar to editing RBR. However, the YFP signal outside ipWER expression region was also hampered by an unknown mechanism, unlike when editing RBR. White arrows mark the neighboring cell walls in a and c. The same construct was used in a and c. Cell walls are highlighted by calcofluor. Numbers indicate the frequency of the observed phenotype in independent T1 samples analyzed. Experiments were repeated three times. Scale bars, 50 μm.
Extended Data Fig. 10 Comparison of IGE system with inducible amiRNA.
a, IGE-PLT2 displays more specific and stronger PLT2-3xYFP downregulation than amiPLT2. After a one-day induction, ip35S»amiPLT2-1 in gPLT2-3xYFP; plt1,2 and ipWOX5»amiPLT2-1 in gPLT2-3xYFP; plt1,2 showed a broader reduction of the YFP signal, particularly in the bracketed regions where no inducible promoter activity was found. Conversely, induced PLT2 editing caused very local loss of the YFP signal. After a three-day induction, the YFP signal is still visible in most of ip35S»amiPLT2-1 in gPLT2-3xYFP; plt1,2 transformants but not in ip35S»Cas9p-PLT2 in gPLT2-3xYFP; plt1,2 transformants. There was no QC differentiation in ipWOX5»amiPLT2-1 in gPLT2-3xYFP; plt1,2 roots. Ctrl refers to 7-day (top panel) or 9-day (bottom panel) old gPLT2-3xYFP; plt1,2. White arrows mark the QC. b, Comparison of the RM (top panel), root secondary growth (middle panel) and cotyledon epidermis (bottom panel) of Col-0, 35S:amiGORBR and ip35S»Cas9p-RBR in Col-0. Inducing RBR editing (germination on 17-β plates for 6 days (top panel), 20 days (middle panel) and 6 days (bottom panel)) resulted in more excessive cell divisions in the LRC than was seen in amiGORBR roots (top panel, germination and six days of growth on 17-β-free plates). Furthermore, RBR editing caused cell overproliferation in phloem (ph) cells and the periderm (pe) of root secondary tissues (middle panel) and pavement cells (pv) and guard cells (gd, blue arrows) of cotyledon epidermis (bottom panel), which was not observed in amiGORBR roots and cotyledons. The knockout (ko) sectors (green dotted line) were frequently accompanied by WT sectors (red dotted line), which can be regarded as an internal control. Red arrows mark guard cells divisions. Cell walls are marked by calcofluor. Numbers indicate the frequency of observed phenotype in independent samples analyzed. Experiments were repeated three times, except experiment on cotyledon epidermis phenotyping, which was repeated two times. Scale bars, 50 μm.
Supplementary information
Supplementary Information
Supplementary methods, Figs. 1–6 and Tables 1–3.
Supplementary Data
TIDE analysis and amplicon deep-sequencing results.
Source data
Source Data Extended Data Fig. 3a
Unprocessed gel.
Source Data Extended Data Fig. 3c
Statistical source data.
Source Data Extended Data Fig. 6b
Statistical source data.
Source Data Supplementary Fig. 4a
Unprocessed gel.
Rights and permissions
About this article
Cite this article
Wang, X., Ye, L., Lyu, M. et al. An inducible genome editing system for plants. Nat. Plants 6, 766–772 (2020). https://doi.org/10.1038/s41477-020-0695-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-020-0695-2
This article is cited by
-
Gibberellins promote polar auxin transport to regulate stem cell fate decisions in cambium
Nature Plants (2023)
-
Cell type-specific mapping of ion distribution in Arabidopsis thaliana roots
Nature Communications (2023)
-
Updates and Applications of CRISPR/Cas Technology in Plants
Journal of Plant Biology (2023)
-
Using CRISPR-Kill for organ specific cell elimination by cleavage of tandem repeats
Nature Communications (2022)
-
General guidelines for CRISPR/Cas-based genome editing in plants
Molecular Biology Reports (2022)