Abstract
Targeted gene editing in hematopoietic stem cells (HSCs) is a promising treatment for several diseases. However, the limited efficiency of homology-directed repair (HDR) in HSCs and the unknown impact of the procedure on clonal composition and dynamics of transplantation have hampered clinical translation. Here, we apply a barcoding strategy to clonal tracking of edited cells (BAR-Seq) and show that editing activates p53, which substantially shrinks the HSC clonal repertoire in hematochimeric mice, although engrafted edited clones preserve multilineage and self-renewing capacity. Transient p53 inhibition restored polyclonal graft composition. We increased HDR efficiency by forcing cell-cycle progression and upregulating components of the HDR machinery through transient expression of the adenovirus 5 E4orf6/7 protein, which recruits the cell-cycle controller E2F on its target genes. Combined E4orf6/7 expression and p53 inhibition resulted in HDR editing efficiencies of up to 50% in the long-term human graft, without perturbing repopulation and self-renewal of edited HSCs. This enhanced protocol should broaden applicability of HSC gene editing and pave its way to clinical translation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All relevant data are included in the manuscript. BAR-Seq and RNA-Seq data are deposited in the Gene Expression Omnibus with the following access codes: GSE143995 (for RNA-Seq) and GSE144340 (BAR-Seq). The reagents described in this manuscript are available under a material transfer agreement with IRCCS Ospedale San Raffaele and Fondazione Telethon; requests for materials should be addressed to L.N.
Code availability
The software for BAR-Seq analysis is freely available at https://bitbucket.org/bereste/bar-seq.
References
Naldini, L. Genetic engineering of hematopoiesis: current stage of clinical translation and future perspectives. EMBO Mol. Med. https://doi.org/10.15252/emmm.201809958 (2019).
Carroll, D. Genome engineering with targetable nucleases. Annu. Rev. Biochem. 83, 409–439 (2014).
Genovese, P. et al. Targeted genome editing in human repopulating haematopoietic stem cells. Nature 510, 235–240 (2014).
Schiroli, G. et al. Preclinical modeling highlights the therapeutic potential of hematopoietic stem cell gene editing for correction of SCID-X1. Sci. Transl. Med. 9, (2017).
Boitano, A. E. et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science 329, 1345–1348 (2010).
Fares, I. et al. Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science 345, 1509–1512 (2014).
Wang, J. et al. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat. Biotechnol. 33, 1256–1263 (2015).
Dever, D. P. et al. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature 539, 384–389 (2016).
De Ravin, S. S. et al. Targeted gene addition in human CD34(+) hematopoietic cells for correction of X-linked chronic granulomatous disease. Nat. Biotechnol. 34, 1–8 (2016).
Kuo, C. Y. et al. Site-specific gene editing of human hematopoietic stem cells for X-linked hyper-IgM syndrome. Cell Rep. https://doi.org/10.1016/j.celrep.2018.04.103 (2018).
Pavel-Dinu, M. et al. Gene correction for SCID-X1 in long-term hematopoietic stem cells. Nat. Commun. https://doi.org/10.1038/s41467-019-09614-y (2019).
Yeh, C. D., Richardson, C. D. & Corn, J. E. Advances in genome editing through control of DNA repair pathways. Nat. Cell Biol. 21, 1468–1478 (2019).
Chu, V. T. et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. https://doi.org/10.1038/nbt.3198 (2015).
Gwiazda, K. S. et al. High efficiency CRISPR/Cas9-mediated gene editing in primary human T-cells using mutant adenoviral E4orf6/E1b55k ‘helper’ proteins. Mol. Ther. 24, 1–11 (2016).
Schiroli, G. et al. Precise gene editing preserves hematopoietic stem cell function following transient p53-Mediated DNA damage response. Cell Stem Cell https://doi.org/10.1016/j.stem.2019.02.019 (2019).
Steensma, D. P. Myelodysplastic syndromes current treatment algorithm 2018. Blood Cancer J. https://doi.org/10.1038/s41408-018-0085-4 (2018).
Lombardo, A. et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat. Methods 8, 861–869 (2011).
Zhao, H., Dahlö, M., Isaksson, A., Syvänen, A. C. & Pettersson, U. The transcriptome of the adenovirus infected cell. Virology https://doi.org/10.1016/j.virol.2011.12.006 (2012).
Täuber, B. & Dobner, T. Adenovirus early E4 genes in viral oncogenesis. Oncogene 20, 7847–7854 (2001).
Seandel, M. et al. Generation of a functional and durable vascular niche by the adenoviral E4ORF1 gene. Proc. Natl Acad. Sci. USA 105, 19288–19293 (2008).
Frese, K. K. et al. Selective PDZ protein-dependent stimulation of phosphatidylinositol 3-kinase by the adenovirus E4-ORF1 oncoprotein. Oncogene https://doi.org/10.1038/sj.onc.1206151 (2003).
Javier, R. T. & Rice, A. P. Emerging theme: cellular PDZ proteins as common targets of pathogenic viruses. J. Virol. https://doi.org/10.1128/JVI.05410-11 (2011).
Huang, M. M. & Hearing, P. The adenovirus early region 4 open reading frame 6/7 protein regulates the DNA binding activity of the cellular transcription factor, E2F, through a direct complex. Genes Dev. 3, 1699–1710 (1989).
Karikó, K., Muramatsu, H., Ludwig, J. & Weissman, D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. https://doi.org/10.1093/nar/gkr695 (2011).
Pattabhi, S. et al. In vivo outcome of homology-directed repair at the HBB Gene in HSC using alternative donor template delivery methods. Mol. Ther. Nucl. Acids https://doi.org/10.1016/j.omtn.2019.05.025 (2019).
Romero, Z. et al. Editing the sickle cell disease mutation in human hematopoietic stem cells: comparison of endonucleases and homologous donor templates. Mol. Ther. https://doi.org/10.1016/j.ymthe.2019.05.014 (2019).
Petrillo, C. et al. Cyclosporine H overcomes innate immune restrictions to improve lentiviral transduction and gene editing in human hematopoietic stem cells. Cell Stem Cell https://doi.org/10.1016/j.stem.2018.10.008 (2018).
Obert, S., O’Connor, R. J., Schmid, S. & Hearing, P. The adenovirus E4-6/7 protein transactivates the E2 promoter by inducing dimerization of a heteromeric E2F complex. Mol. Cell. Biol. 14, 1333–1346 (1994).
Menendez, D., Nguyen, T. A., Snipe, J. & Resnick, M. A. The cytidine deaminase APOBEC3 family is subject to transcriptional regulation by p53. Mol. Cancer Res. https://doi.org/10.1158/1541-7786.MCR-17-0019 (2017).
Aleem, E., Kiyokawa, H. & Kaldis, P. Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat. Cell Biol. https://doi.org/10.1038/ncb1284 (2005).
Radhakrishnan, S. K. et al. Constitutive expression of E2F-1 leads to p21-dependent cell cycle arrest in S phase of the cell cycle. Oncogene https://doi.org/10.1038/sj.onc.1207571 (2004).
Komori, H., Enomoto, M., Nakamura, M., Iwanaga, R. & Ohtani, K. Distinct E2F-mediated transcriptional program regulates p14ARF gene expression. EMBO J. https://doi.org/10.1038/sj.emboj.7600836 (2005).
Querido, E. et al. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15, 3104–3117 (2001).
Mjelle, R. et al. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA Repair https://doi.org/10.1016/j.dnarep.2015.03.007 (2015).
Clement, K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nature Biotech. https://doi.org/10.1038/s41587-019-0032-3 (2019).
Milyavsky, M. et al. A Distinctive DNA damage response in human hematopoietic stem cells reveals an apoptosis-independent role for p53 in self-renewal. Cell Stem Cell 7, 186–197 (2010).
van den Berg, J. et al. A limited number of double-strand DNA breaks is sufficient to delay cell cycle progression. Nucleic Acids Res. https://doi.org/10.1093/nar/gky786 (2018).
Wang, J. C. Y., Doedens, M. & Dick, J. E. Primitive human hematopoietic cells are enriched in cord blood compared with adult bone marrow or mobilized peripheral blood as measured by the quantitative in vivo SCID-repopulating cell assay. Blood https://doi.org/10.1182/blood.v89.11.3919 (1997).
Zonari, E. et al. Efficient ex vivo engineering and expansion of highly purified human hematopoietic stem and progenitor cell populations for gene therapy. Stem Cell Rep. 8, 977–990 (2017).
Wagenblast, E. et al. Functional profiling of single CRISPR/Cas9-edited human long-term hematopoietic stem cells. Nat. Commun. https://doi.org/10.1038/s41467-019-12726-0 (2019).
Bai, T. et al. Expansion of primitive human hematopoietic stem cells by culture in a zwitterionic hydrogel. Nat. Med. https://doi.org/10.1038/s41591-019-0601-5 (2019).
Hoban, M. D. et al. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood https://doi.org/10.1182/blood-2014-12-615948 (2015).
Beerman, I., Seita, J., Inlay, M. A., Weissman, I. L. & Rossi, D. J. Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell https://doi.org/10.1016/j.stem.2014.04.016 (2014).
Laurenti, E. et al. CDK6 levels regulate quiescence exit in human hematopoietic stem cells. Cell Stem Cell https://doi.org/10.1016/j.stem.2015.01.017 (2015).
Schaley, J., O’Connor, R. J., Taylor, L. J., Bar-Sagi, D. & Hearing, P. Induction of the cellular E2F-1 promoter by the adenovirus E4-6/7 protein. J. Virol. https://doi.org/10.1128/jvi.74.5.2084-2093.2000 (2000).
Schaley, J. E., Polonskaia, M. & Hearing, P. The adenovirus E4-6/7 protein directs nuclear localization of E2F-4 via an Arginine-Rich Motif. J. Virol. https://doi.org/10.1128/jvi.79.4.2301-2308.2005 (2005).
Stanelle, J., Stiewe, T., Theseling, C. C., Peter, M. & Pützer, B. M. Gene expression changes in response to E2F1 activation. Nucleic Acids Res. https://doi.org/10.1093/nar/30.8.1859 (2002).
Yin, A. H. et al. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood 90, 5002–5012 (1997).
Notta, F. et al. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science https://doi.org/10.1126/science.1201219 (2011).
Fares, I. et al. EPCR expression marks UM171-expanded CD34 + cord blood stem cells. Blood https://doi.org/10.1182/blood-2016-11-750729 (2017).
Haapaniemi, E., Botla, S., Persson, J., Schmierer, B. & Taipale, J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. https://doi.org/10.1038/s41591-018-0049-z (2018).
Ihry, R. J. et al. P53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. https://doi.org/10.1038/s41591-018-0050-6 (2018).
Sakofsky, C. J. et al. Repair of multiple simultaneous double-strand breaks causes bursts of genome-wide clustered hypermutation. PLoS Biol. https://doi.org/10.1371/journal.pbio.3000464 (2019).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. https://doi.org/10.1038/nbt.2647 (2013).
Notredame, C., Higgins, D. G. & Heringa, J. T-coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. https://doi.org/10.1006/jmbi.2000.4042 (2000).
Provasi, E. et al. Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nat. Med. https://doi.org/10.1038/nm.2700 (2012).
Lassmann, T. TagDust2: a generic method to extract reads from sequencing data. BMC Bioinformatics https://doi.org/10.1186/s12859-015-0454-y (2015).
Del Core, L., Montini, E., Di Serio, C. & Calabria, A. Dealing with data evolution and data integration: an approach using rarefaction. in Proc. 49th Scientific Meeting of the Italian Statistical Society https://meetings3.sis-statistica.org/index.php/sis2018/49th/paper/view/1511 (2018).
Pinheiro, J. & Bates, D. Linear and nonlinear mixed effects models (nlme). R package version 3.1-318 (2018).
Bates, D., Mächler, M., Bolker, B. M. & Walker, S. C. Fitting linear mixed-effects models using lme4. J. Stat. Softw. https://doi.org/10.18637/jss.v067.i01 (2015).
Acknowledgements
We thank all members of L.N.’s laboratory for discussion, the IRCCS San Raffaele Hospital Flow Cytometry facility, the IRCCS San Raffaele Center for Omics Sciences (COSR), A. Auricchio and M. Doria (Telethon Institute of GEnetics and Medicine; TIGEM Vector Core, Pozzuoli, Italy) for providing AAV6 vectors, E. Ayuso (INSERM UMR1089, Nantes, France) for providing comments on AAV biology, L. Periè (Institute Curie, Paris, France) and J. Urbanus (the Netherlands Cancer Institute, Amsterdam, the Netherlands) for advice on the BAR cloning strategy, G. Schiroli for initial help with the design of the BAR-Seq strategy, R. Di Micco and B. Gentner for critical reading of the manuscript. We thank T. Plati for technical support in ddPCR analyses, T. Di Tomaso and G. Desantis for purifying mPB HSPCs, F. Benedicenti for helping in library preparation for NHEJ clonal tracking, L. Sergi Sergi, I. Cuccovillo, M. Biffi and M. Soldi for IDLV production and purification (SR-TIGET, IRCCS San Raffaele Scientific Institute), C. Di Serio for coordinating CUSSB support (Vita-Salute San Raffaele University). This work was supported by grants to: L.N. from Telethon (TIGET grant no. E4), the Italian Ministry of Health (grant nos. PE-2016-02363691; E-Rare-3 JTC 2017), the Italian Ministry of University and Research (PRIN 2017 Prot. no. 20175XHBPN), the EU Horizon 2020 Program (UPGRADE) and from the Louis-Jeantet Foundation through the 2019 Jeantet-Collen Prize for Translational Medicine; P.G. from Telethon (TIGET grant no. E3) and the Italian Ministry of Health (grant no. GR-2013-02358956); A.K.R. from the ERC (ImmunoStem, grant no. 819815). S.F., V.V. and G.U. conducted this study as partial fulfillment of their PhD in Molecular Medicine, International PhD School, Vita-Salute San Raffaele University (Milan, Italy). A.J. conducted this study as partial fulfillment of his PhD in Translational and Molecular Medicine DIMET, Milano-Bicocca University (Monza, Italy) with M. Serafini acting as the university tutor.
Author information
Authors and Affiliations
Contributions
S.F. and A.J. performed research, interpreted data and wrote the manuscript. G.U. performed experiments with IDLV-based repair template supervised by A.K.R. L.A. provided technical help with mouse experiments. V.V. performed CD40LG and T cell experiments. S.B., D.C., D.L. and I.M. performed bioinformatic analysis. C.B. and F.C. performed statistical analyses. P.G. and L.N. designed the study, interpreted data, supervised research and wrote the manuscript. L.N. coordinated the work.
Corresponding author
Ethics declarations
Competing interests
L.N., P.G. and A.K.R. are inventors of patents on applications of gene editing in HSPCs owned and managed by the San Raffaele Scientific Institute and the Telethon Foundation, including a patent application on improved gene editing filed by S.F., A.J., P.G. and L.N. L.N. is founder and quota holder and P.G. is quota holder of GeneSpire, a startup company aiming to develop ex vivo gene editing in genetic diseases. All other authors declare no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 BAR-Seq dissects clonal dynamics of HDR-edited cells.
a, Percentage of GFP+ cells within subpopulations 96 h after AAVS1 editing with the barcoded or non-barcoded AAV6 (3 HSPC donors; n = 4). Median. b, Number of unique BARs and relative abundances in bulk cultured HSPCs 72 h after editing. One representative sample out of two is shown. c, Experimental scheme. d, e, Culture composition (d) and percentage of GFP+ cells within subpopulations (e) of AAVS1 edited HSPCs with the indicated treatments at the time of transplant and 96 h after editing, respectively (10 HSPC donors; n = 1). f, g, Percentage of hCD45+ cells (f) and GFP+ cells within human graft (g) in BM or spleen (SPL) of mice from Fig. 1c (n = 9, 10, 6, 3). Median. Kruskal-Wallis test. h, i, Abundance of ranked BARs from PBMCs collected at 8 (h) and 12 (i) weeks after transplant, as in Fig. 1e. j, Heatmap as in Fig. 1f for ‘w/o S/U (+4 days)’-transplanted mice. k, Number of dominant unique BARs in sorted hCD45+ cell lineages and HSPCs of mice from Fig. 1c. Mice with % of circulating hCD45+GFP+ cells at 18 weeks timepoints < 0.1 were plotted with BAR count = 0 (n = 9, 10, 10, 10). Median. l, Correlation between the percentage of GFP+ cells (within hCD45+) and the number of dominant unique BARs in “w/o S/U”, “RNP + AAV6” and “+ GSE56” mice of this study (n = 71). Each dot represents one mouse. Mice with number of dominant unique BARs ≥6 (arbitrary threshold) are shown in magenta (coefficient of variation (CV) = 0.51); mice with number of dominant unique BARs <5 are shown in yellow (CV = 0.87). Dashed line indicates the median percentage of GFP+ cells within CD90+ HSPCs in the in vitro outgrown of transplanted edited cells. m, Longitudinal PBMC analysis as in Fig. 1k but including in the analysis >95% of total BAR reads (n = 4, 5). Median. n, Correlation as in Fig. 1l at 8 weeks after transplant (n = 28). Spearman correlation coefficient was calculated. All statistical tests are two-tailed. n indicate independent animals.
Extended Data Fig. 2 Identification of Ad protein variants improving HDR efficiency.
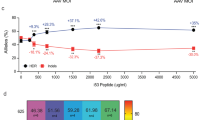
a, b, Multiple sequences alignment of E4orf1 (a) and E4orf6/7 (b) Ad variants. Sequences were collected from online RCSB Protein Data Bank and UniProt. c, d, Percentage of HDR-edited alleles (c) and GFP+ cells within subpopulations (d) 96 h after AAVS1 editing in bulk CB HSPCs with indicated treatments (n = 4, 4, 4, 4; other treatments: n = 2). Median. e, FACS plots of untreated (UT) and AAV6-transduced HSPCs in absence (“Mock AAV6”) or presence of Ad5-E1B55K+Ad5-E4orf6 measured 24 h after treatments. The results of one representative experiment out of three is shown. f, Number of colonies from bulk edited HSPCs in the indicated treatments (n = 2). Mean. g, h, Fold change expansion of live HSPCs after indicated treatments from Extended Data Fig. 2c (n = 2). Median. i, Number of colonies from bulk edited HSPCs with the indicated treatments (n = 2). Mean. j, CD90 MFI in edited HSPCs measured 96 h after editing with indicated treatments (n = 6). Median with IQR. Friedman test with two-tailed Dunn’s multiple comparisons. k, Percentage of live, early/late apoptotic and necrotic bulk HSPCs 24 h after editing with the indicated treatments (7 HSPC donors; n = 3). Mean ± SEM. l-m, Percentage of HDR/NHEJ-edited alleles (l) and culture composition (m) 96 h after editing of bulk mPB HSPCs from Fig. 2h (n = 3). Mean ± SEM. n) Percentage of GFP+ T cells 14 days after AAVS1 editing with indicated treatments (n = 3). Median. o-p, Percentage of HDR/NHEJ-edited alleles (o) and culture composition (p) 96 h after IDLV-based editing of bulk CB HSPCs from Fig. 2i (n = 3). Mean ± SEM. Red arrows indicate Ad protein variants selected for further investigation. n indicate independent experiments.
Extended Data Fig. 3 Investigating the transcriptional response upon enhanced editing.
a, b, Fold change expression of cell cycle related genes relative to UT 24 h after AAV-based editing with the indicated treatments in CB (a) or mPB (b) HSPCs (CB: n = 8, 5, 7, 6, 3, 3, 3, 3; mPB: n = 4, 4, 4, 3). Median. c, Fold change expression of CDKN1A relative to UT 24 h after IDLV-based editing with indicated treatments in CB HSPCs (n = 3). Median. d, Fold change expression of CDKN1A relative to UT at 24 h after AAV-based editing with indicated treatments in CB HSPCs (n = 5). Median. e) MA plots showing significant down- (green) and up- (red) regulated genes after AAVS1 editing in mock electroporated (left) and standard edited (right) compared to UT (n = 3). PPP1R12C, the AAVS1 hosting gene appears among the downregulated genes, concordantly with previous reports showing transient transcriptional repression at the site of DNA DSB15. f) Random walk plots for the indicated Reactome categories. Relative adjusted p-values and NES are shown. g) Venn diagram showing the number of genes related to the “Allograft rejection” category upregulated upon standard editing and downregulated in presence of “+ Ad5-E4orf6/7” treatment. h) Venn diagram showing the number of HDR genes (“Homology directed repair” category from Reactome database) shared with E2F pathway target genes (Hallmark gene set) from cluster 1 or other clusters from Fig. 3e. i) Schematic of “cell cycle” and “p53 pathway” KEGG gene ontologies highlighting genes (red) belonging to clusters 1 (top) and 3 (bottom) of Fig. 3e. For all panels with statistical analysis: Friedman test with two-tailed Dunn’s multiple comparisons. n indicate independent experiments, except for Extended Data Fig. 3e where n indicates independent samples.
Extended Data Fig. 4 Transplantation of enhancer-edited HSPCs in NSG mice.
a, Experimental workflow. b, Percentage of hCD45+ cells in SPL and BM of mice from Figs. 4a, b (n = 23, 11, 15, 16). LME followed by post-hoc analysis. Mean ± SEM. c, BM cell composition in mice from Fig. 4a, b. LME followed by post-hoc analysis for HSPCs (n = 23, 11, 15, 16). Mean ± SEM. d, Percentage of cells harboring monoallelic or biallelic integration(s) in SPL of mice from Fig. 4a, b (n = 23, 11, 15, 16). Mean ± SEM. e, Percentage of circulating hCD45+ cells in mice transplanted with CB HSPCs IL2RG-edited in presence of GSE56 and Ad5 E4orf6/7 (n = 4). Comparison with the previously published results for “RNP+AAV6” and “+ GSE56” groups22 is shown (n = 5, 6). All statistical tests are two-tailed. n indicate independent animals.
Extended Data Fig. 5 Enhanced editing preserves multilineage repopulation capacity and self-renewing potential of individual edited HSPC clones.
a, Heatmap showing the abundance (red-scaled palette) of dominant unique BARs (rows) retrieved in PBMCs at indicated times after transplant and sorted hCD45+ cell lineages of mice from one experiment of Fig. 4a (separated columns). b, Clonal diversity within sorted hCD45+ cell lineages in mice from Extended Data Fig. 5a (B cells: n = 5, 3, 5; Myeloid and T cells: n = 6, 3, 3). Median. Two-tailed Friedman test with Dunn’s multiple comparisons. Experimental groups were unified for statistical analysis. c, Number of dominant unique BARs in PBMCs or BM of mice from one experiment in Fig. 4a (PBMCs: n = 5, 4; BM: n = 3, 3). Median. d, Percentage of NHEJ-edited alleles within the non-HDR edited fraction from Fig. 5g (n = 6, 3, 6). Median. e, Heatmaps as in Extended Data Fig. 5a showing the dominant unique BARs in 9-weeks PBMCs and in sorted hCD45+ cell lineages (15 weeks) of secondary recipients. n indicate independent animals.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2.
Supplementary Tables 1–4
This file includes the following items. Supplementary Table 1, complete gene list from Fig. 3e. Supplementary Table 2, complete gene list and fold change expression from Fig. 3f. Supplementary Table 3, list of primers and probes. Supplementary Table 4, details on mRNA-expressing vectors and the final concentration used.
Supplementary Table 5
Detailed statistical analyses.
Rights and permissions
About this article
Cite this article
Ferrari, S., Jacob, A., Beretta, S. et al. Efficient gene editing of human long-term hematopoietic stem cells validated by clonal tracking. Nat Biotechnol 38, 1298–1308 (2020). https://doi.org/10.1038/s41587-020-0551-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-020-0551-y
This article is cited by
-
Precise genome-editing in human diseases: mechanisms, strategies and applications
Signal Transduction and Targeted Therapy (2024)
-
Transient inhibition of 53BP1 increases the frequency of targeted integration in human hematopoietic stem and progenitor cells
Nature Communications (2024)
-
A potential paradigm in CRISPR/Cas systems delivery: at the crossroad of microalgal gene editing and algal-mediated nanoparticles
Journal of Nanobiotechnology (2023)
-
Genotoxic effects of base and prime editing in human hematopoietic stem cells
Nature Biotechnology (2023)
-
CRISPR-Cas9 engineering of the RAG2 locus via complete coding sequence replacement for therapeutic applications
Nature Communications (2023)