Abstract
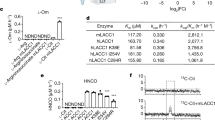
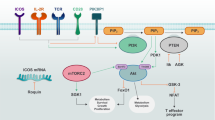
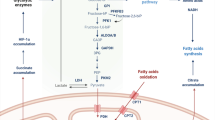
Immunometabolism plays a fundamental role in health and diseases and involves multiple genes and signals. Aconitate decarboxylase 1 (ACOD1; also known as IRG1) is emerging as a regulator of immunometabolism in inflammation and infection. Upregulation of ACOD1 expression occurs in activated immune cells (e.g., macrophages and monocytes) in response to pathogen infection (e.g., bacteria and viruses), pathogen-associated molecular pattern molecules (e.g., LPS), cytokines (e.g., TNF and IFNs), and damage-associated molecular patterns (e.g., monosodium urate). Mechanistically, several immune receptors (e.g., TLRs and IFNAR), adapter proteins (e.g., MYD88), ubiquitin ligases (e.g., A20), and transcription factors (e.g., NF-κB, IRFs, and STATs) form complex signal transduction networks to control ACOD1 expression in a context-dependent manner. Functionally, ACOD1 mediates itaconate production, oxidative stress, and antigen processing and plays dual roles in immunity and diseases. On the one hand, activation of the ACOD1 pathway may limit pathogen infection and promote embryo implantation. On the other hand, abnormal ACOD1 expression can lead to tumor progression, neurodegenerative disease, and immune paralysis. Further understanding of the function and regulation of ACOD1 is important for the application of ACOD1-based therapeutic strategies in disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Mogensen, T. H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol Rev. 22, 240–273 (2009).
Akira, S., Uematsu, S. & Takeuchi, O. Pathogen recognition and innate immunity. Cell 124, 783–801 (2006).
Chaplin, D. D. Overview of the immune response. J. Allergy Clin. Immunol. 125, S3–S23 (2010).
O’Neill, L. A., Kishton, R. J. & Rathmell, J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 16, 553–565 (2016).
Mathis, D. & Shoelson, S. E. Immunometabolism: an emerging frontier. Nat. Rev. Immunol. 11, 81 (2011).
Michelucci, A. et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl Acad. Sci. USA 110, 7820–7825 (2013).
Cordes, T. et al. Immunoresponsive gene 1 and itaconate inhibit succinate dehydrogenase to modulate intracellular succinate levels. J. Biol. Chem. 291, 14274–14284 (2016).
Lee, C. G., Jenkins, N. A., Gilbert, D. J., Copeland, N. G. & O’Brien, W. E. Cloning and analysis of gene regulation of a novel LPS-inducible cDNA. Immunogenetics 41, 263–270 (1995).
Shi, S. et al. Expression of many immunologically important genes in Mycobacterium tuberculosis-infected macrophages is independent of both TLR2 and TLR4 but dependent on IFN-alphabeta receptor and STAT1. J. Immunol. 175, 3318–3328 (2005).
Hoshino, K., Kaisho, T., Iwabe, T., Takeuchi, O. & Akira, S. Differential involvement of IFN-beta in Toll-like receptor-stimulated dendritic cell activation. Int Immunol. 14, 1225–1231 (2002).
Daniels, B. P. et al. The nucleotide sensor ZBP1 and kinase RIPK3 induce the enzyme IRG1 to promote an antiviral metabolic state in neurons. Immunity 50, 64–76. e64 (2019).
Dix, A. et al. Biomarker-based classification of bacterial and fungal whole-blood infections in a genome-wide expression study. Front. Microbiol. 6, 171 (2015).
Chen, B., Zhang, D. & Pollard, J. W. Progesterone regulation of the mammalian ortholog of methylcitrate dehydratase (immune response gene 1) in the uterine epithelium during implantation through the protein kinase C pathway. Mol. Endocrinol. 17, 2340–2354 (2003).
Degrandi, D., Hoffmann, R., Beuter-Gunia, C. & Pfeffer, K. The proinflammatory cytokine-induced IRG1 protein associates with mitochondria. J. Interferon Cytokine Res. 29, 55–67 (2009).
Dominguez-Andres, J. et al. The itaconate pathway is a central regulatory node linking innate immune tolerance and trained immunity. Cell Metab. 29, 211–220. e215 (2019).
Hall, C. J. et al. Immunoresponsive gene 1 augments bactericidal activity of macrophage-lineage cells by regulating beta-oxidation-dependent mitochondrial ROS production. Cell Metab. 18, 265–278 (2013).
Pan, J. et al. Immune responsive gene 1, a novel oncogene, increases the growth and tumorigenicity of glioma. Oncol. Rep. 32, 1957–1966 (2014).
Cheon, Y. P., Xu, X., Bagchi, M. K. & Bagchi, I. C. Immune-responsive gene 1 is a novel target of progesterone receptor and plays a critical role during implantation in the mouse. Endocrinology 144, 5623–5630 (2003).
Li, H. et al. Different neurotropic pathogens elicit neurotoxic CCR9- or neurosupportive CXCR3-expressing microglia. J. Immunol. 177, 3644–3656 (2006).
Li, Y. et al. Immune responsive gene 1 (IRG1) promotes endotoxin tolerance by increasing A20 expression in macrophages through reactive oxygen species. J. Biol. Chem. 288, 16225–16234 (2013).
Lampropoulou, V. et al. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab. 24, 158–166 (2016).
Yin, S. et al. The IRG1-itaconate axis promotes viral replication via metabolic reprogramming and protein prenylation. Cell Metab. 19-00910, 54 (2019).
Brubaker, S. W., Bonham, K. S., Zanoni, I. & Kagan, J. C. Innate immune pattern recognition: a cell biological perspective. Annu. Rev. Immunol. 33, 257–290 (2015).
Medzhitov, R. & Janeway, C. Jr. Innate immunity. N. Engl. J. Med. 343, 338–344 (2000).
Xiao, W. et al. Expression profile of human immune-responsive gene 1 and generation and characterization of polyclonal antiserum. Mol. Cell Biochem. 353, 177–187 (2011).
Schmidt, G. & Richter, K. Expression pattern of XIRG, a marker for non-neural ectoderm. Dev. Genes Evol. 210, 575–578 (2000).
Chen, F. et al. Crystal structure of cis-aconitate decarboxylase reveals the impact of naturally occurring human mutations on itaconate synthesis. Proc. Natl Acad. Sci. USA 116, 20644–20654 (2019).
Naujoks, J. et al. IFNs modify the proteome of legionella-containing vacuoles and restrict infection via IRG1-derived itaconic acid. PLoS Pathog. 12, e1005408 (2016).
Kim, Y. J. et al. Botulinum neurotoxin type A induces TLR2-mediated inflammatory responses in macrophages. PLoS ONE 10, e0120840 (2015).
Mills, E. L. et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556, 113–117 (2018).
Kim, J. Y. et al. Radioprotective effect of newly synthesized toll-like receptor 5 agonist, KMRC011, in mice exposed to total-body irradiation. J. Radiat. Res. 60, 432–441 (2019).
Kong, F. et al. Transcriptional profiling in experimental visceral leishmaniasis reveals a broad splenic inflammatory environment that conditions macrophages toward a disease-promoting phenotype. PLoS Pathog. 13, e1006165 (2017).
Sherwin, J. R. et al. Identification of genes regulated by leukemia-inhibitory factor in the mouse uterus at the time of implantation. Mol. Endocrinol. 18, 2185–2195 (2004).
Catalano, R. D. et al. Inhibition of Stat3 activation in the endometrium prevents implantation: a nonsteroidal approach to contraception. Proc. Natl Acad. Sci. USA 102, 8585–8590 (2005).
Terakawa, J. et al. Embryo implantation is blocked by intraperitoneal injection with anti-LIF antibody in mice. J. Reprod. Dev. 57, 700–707 (2011).
Cheon, Y. P. et al. A genomic approach to identify novel progesterone receptor regulated pathways in the uterus during implantation. Mol. Endocrinol. 16, 2853–2871 (2002).
Pessler, F. et al. Identification of novel monosodium urate crystal regulated mRNAs by transcript profiling of dissected murine air pouch membranes. Arthritis Res. Ther. 10, R64 (2008).
Jamal Uddin, M. et al. IRG1 induced by heme oxygenase-1/carbon monoxide inhibits LPS-mediated sepsis and pro-inflammatory cytokine production. Cell Mol. Immunol. 13, 170–179 (2016).
Kane, M. J. et al. Altered gene expression in cultured microglia in response to simulated blast overpressure: possible role of pulse duration. Neurosci. Lett. 522, 47–51 (2012).
Basler, T., Jeckstadt, S., Valentin-Weigand, P. & Goethe, R. Mycobacterium paratuberculosis, Mycobacterium smegmatis, and lipopolysaccharide induce different transcriptional and post-transcriptional regulation of the IRG1 gene in murine macrophages. J. Leukoc. Biol. 79, 628–638 (2006).
Papathanassiu, A. E. et al. BCAT1 controls metabolic reprogramming in activated human macrophages and is associated with inflammatory diseases. Nat. Commun. 8, 16040 (2017).
Gonzalez-Pena, D. et al. Differential transcriptome networks between IDO1-knockout and wild-type mice in brain microglia and macrophages. PLoS ONE 11, e0157727 (2016).
Munn, D. H. & Mellor, A. L. Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol. 34, 137–143 (2013).
Kawai, T. et al. Lipopolysaccharide stimulates the MyD88-independent pathway and results in activation of IFN-regulatory factor 3 and the expression of a subset of lipopolysaccharide-inducible genes. J. Immunol. 167, 5887–5894 (2001).
Wei, S. et al. Overexpression of Toll-like receptor 4 enhances LPS-induced inflammatory response and inhibits Salmonella Typhimurium growth in ovine macrophages. Eur. J. Cell Biol. 98, 36–50 (2019).
Rodriguez, N. et al. MyD88-dependent changes in the pulmonary transcriptome after infection with Chlamydia pneumoniae. Physiol. Genom. 30, 134–145 (2007).
Zhao, G. N., Jiang, D. S. & Li, H. Interferon regulatory factors: at the crossroads of immunity, metabolism, and disease. Biochim. Biophys. Acta 1852, 365–378 (2015).
Tangsudjai, S. et al. Involvement of the MyD88-independent pathway in controlling the intracellular fate of Burkholderia pseudomallei infection in the mouse macrophage cell line RAW 264.7. Microbiol. Immunol. 54, 282–290 (2010).
Ganta, V. C. et al. A MicroRNA93-interferon regulatory factor-9-immunoresponsive gene-1-itaconic acid pathway modulates M2-like macrophage polarization to revascularize ischemic muscle. Circulation. 135, 2403–2425 (2017).
Tallam, A. et al. Gene regulatory network inference of immunoresponsive gene 1 (IRG1) identifies interferon regulatory factor 1 (IRF1) as its transcriptional regulator in mammalian macrophages. PLoS ONE 11, e0149050 (2016).
Opipari, A. W. Jr., Boguski, M. S. & Dixit, V. M. The A20 cDNA induced by tumor necrosis factor alpha encodes a novel type of zinc finger protein. J. Biol. Chem. 265, 14705–14708 (1990).
Jaattela, M., Mouritzen, H., Elling, F. & Bastholm, L. A20 zinc finger protein inhibits TNF and IL-1 signaling. J. Immunol. 156, 1166–1173 (1996).
Van Quickelberghe, E. et al. Identification of immune-responsive gene 1 (IRG1) as a target of A20. J. Proteome Res. 17, 2182–2191 (2018).
Newton, A. C. Protein kinase C: structure, function, and regulation. J. Biol. Chem. 270, 28495–28498 (1995).
Konishi, H. et al. Activation of protein kinase C by tyrosine phosphorylation in response to H2O2. Proc. Natl Acad. Sci. USA 94, 11233–11237 (1997).
Aronoff, D. M., Canetti, C., Serezani, C. H., Luo, M. & Peters-Golden, M. Cutting edge: macrophage inhibition by cyclic AMP (cAMP): differential roles of protein kinase A and exchange protein directly activated by cAMP-1. J. Immunol. 174, 595–599 (2005).
Hanada, T. & Yoshimura, A. Regulation of cytokine signaling and inflammation. Cytokine Growth Factor Rev. 13, 413–421 (2002).
Day, D. A. & Tuite, M. F. Post-transcriptional gene regulatory mechanisms in eukaryotes: an overview. J. Endocrinol. 157, 361–371 (1998).
Filipowicz, W., Bhattacharyya, S. N. & Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9, 102–114 (2008).
Shi, H. Z., Wang, D., Sun, X. N. & Sheng, L. MicroRNA-378 acts as a prognosis marker and inhibits cell migration, invasion and epithelial-mesenchymal transition in human glioma by targeting IRG1. Eur. Rev. Med Pharmacol. Sci. 22, 3837–3846 (2018).
Kelly, B. & O’Neill, L. A. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 25, 771–784 (2015).
Pearce, E. L. & Pearce, E. J. Metabolic pathways in immune cell activation and quiescence. Immunity 38, 633–643 (2013).
Lohkamp, B., Bauerle, B., Rieger, P. G. & Schneider, G. Three-dimensional structure of iminodisuccinate epimerase defines the fold of the MmgE/PrpD protein family. J. Mol. Biol. 362, 555–566 (2006).
Strelko, C. L. et al. Itaconic acid is a mammalian metabolite induced during macrophage activation. J. Am. Chem. Soc. 133, 16386–16389 (2011).
Shin, J. H. et al. (1)H NMR-based metabolomic profiling in mice infected with Mycobacterium tuberculosis. J. Proteome Res. 10, 2238–2247 (2011).
Kobayashi, A. et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell Biol. 24, 7130–7139 (2004).
Bambouskova, M. et al. Electrophilic properties of itaconate and derivatives regulate the IkappaBzeta-ATF3 inflammatory axis. Nature 556, 501–504 (2018).
Ahmed, S. M., Luo, L., Namani, A., Wang, X. J. & Tang, X. Nrf2 signaling pathway: Pivotal roles in inflammation. Biochim. Biophys. Acta Mol. Basis Dis. 1863, 585–597 (2017).
Yi, Z. et al. IRG1/Itaconate activates Nrf2 in hepatocytes to protect against liver ischemia-reperfusion injury. Hepatology. https://doi.org/10.1002/hep.31147 (2020).
Kornberg, M. D. et al. Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science 360, 449–453 (2018).
Liao, S. T. et al. 4-Octyl itaconate inhibits aerobic glycolysis by targeting GAPDH to exert anti-inflammatory effects. Nat. Commun. 10, 5091 (2019).
Mills, E. L. et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell 167, 457–470. e413 (2016).
Tannahill, G. M. et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature 496, 238–242 (2013).
Ackermann, W. W. & Potter, V. R. Enzyme inhibition in relation to chemotherapy. Proc. Soc. Exp. Biol. Med. 72, 1–9 (1949).
Nemeth, B. et al. Abolition of mitochondrial substrate-level phosphorylation by itaconic acid produced by LPS-induced Irg1 expression in cells of murine macrophage lineage. FASEB J. 30, 286–300 (2016).
Ahn, S., Jung, J., Jang, I. A., Madsen, E. L. & Park, W. Role of glyoxylate shunt in oxidative stress response. J. Biol. Chem. 291, 11928–11938 (2016).
Palsuledesai, C. C. & Distefano, M. D. Protein prenylation: enzymes, therapeutics, and biotechnology applications. ACS Chem. Biol. 10, 51–62 (2015).
Einav, S. & Glenn, J. S. Prenylation inhibitors: a novel class of antiviral agents. J. Antimicrob. Chemother. 52, 883–886 (2003).
Liu, Y. et al. N (6)-methyladenosine RNA modification-mediated cellular metabolism rewiring inhibits viral replication. Science 365, 1171–1176 (2019).
Alfadda, A. A. & Sallam, R. M. Reactive oxygen species in health and disease. J. Biomed. Biotechnol. 2012, 936486 (2012).
Paiva, C. N. & Bozza, M. T. Are reactive oxygen species always detrimental to pathogens? Antioxid. Redox Signal. 20, 1000–1037 (2014).
Liu, X., Wu, X. P., Zhu, X. L., Li, T. & Liu, Y. IRG1 increases MHC class I level in macrophages through STAT-TAP1 axis depending on NADPH oxidase mediated reactive oxygen species. Int. Immunopharmacol. 48, 76–83 (2017).
West, A. P. et al. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature 472, 476–480 (2011).
Bergsbaken, T., Fink, S. L. & Cookson, B. T. Pyroptosis: host cell death and inflammation. Nat. Rev. Microbiol 7, 99–109 (2009).
Rossjohn, J. et al. T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33, 169–200 (2015).
Hewitt, E. W. The MHC class I antigen presentation pathway: strategies for viral immune evasion. Immunology 110, 163–169 (2003).
Ackerman, A. L. & Cresswell, P. Cellular mechanisms governing cross-presentation of exogenous antigens. Nat. Immunol. 5, 678–684 (2004).
Liu, X. et al. Polymorphisms in IRG1 gene associated with immune responses to hepatitis B vaccination in a Chinese Han population and function to restrain the HBV life cycle. J. Med. Virol. 89, 1215–1223 (2017).
Smith, J. et al. Systems analysis of immune responses in Marek’s disease virus-infected chickens identifies a gene involved in susceptibility and highlights a possible novel pathogenicity mechanism. J. Virol. 85, 11146–11158 (2011).
Gautam, A. et al. Interleukin-10 alters effector functions of multiple genes induced by Borrelia burgdorferi in macrophages to regulate Lyme disease inflammation. Infect. Immun. 79, 4876–4892 (2011).
Nair, S. et al. Irg1 expression in myeloid cells prevents immunopathology during M. tuberculosis infection. J. Exp. Med. 215, 1035–1045 (2018).
Cho, H. et al. Differential innate immune response programs in neuronal subtypes determine susceptibility to infection in the brain by positive-stranded RNA viruses. Nat. Med. 19, 458–464 (2013).
Ren, K. et al. Suppression of IRG-1 reduces inflammatory cell infiltration and lung injury in respiratory syncytial virus infection by reducing production of reactive oxygen species. J. Virol. 90, 7313–7322 (2016).
McNeal, S. et al. Association of immunosuppression with DR6 expression during the development and progression of spontaneous ovarian cancer in Laying Hen model. J. Immunol. Res 2016, 6729379 (2016).
Weiss, J. M. et al. Itaconic acid mediates crosstalk between macrophage metabolism and peritoneal tumors. J. Clin. Investig 128, 3794–3805 (2018).
Fischer, C. et al. Bisphenol A (BPA) exposure in utero leads to immunoregulatory cytokine dysregulation in the mouse mammary gland: a potential mechanism programming breast cancer risk. Horm. Cancer 7, 241–251 (2016).
Perry, V. H. & Teeling, J. Microglia and macrophages of the central nervous system: the contribution of microglia priming and systemic inflammation to chronic neurodegeneration. Semin Immunopathol. 35, 601–612 (2013).
Duffy, C. M., Hofmeister, J. J., Nixon, J. P. & Butterick, T. A. High fat diet increases cognitive decline and neuroinflammation in a model of orexin loss. Neurobiol. Learn Mem. 157, 41–47 (2019).
Mor, G., Cardenas, I., Abrahams, V. & Guller, S. Inflammation and pregnancy: the role of the immune system at the implantation site. Ann. N. Y. Acad. Sci. 1221, 80–87 (2011).
Paria, B. C., Huet-Hudson, Y. M. & Dey, S. K. Blastocyst’s state of activity determines the “window” of implantation in the receptive mouse uterus. Proc. Natl Acad. Sci. USA 90, 10159–10162 (1993).
Salleh, N. & Giribabu, N. Leukemia inhibitory factor: roles in embryo implantation and in nonhormonal contraception. ScientificWorldJournal 2014, 201514 (2014).
Matulova, M. et al. Characterization of chicken spleen transcriptome after infection with Salmonella enterica serovar enteritidis. PLoS ONE 7, e48101 (2012).
Matulova, M. et al. Chicken innate immune response to oral infection with Salmonella enterica serovar enteritidis. Vet. Res. 44, 37 (2013).
Rychlik, I., Elsheimer-Matulova, M. & Kyrova, K. Gene expression in the chicken caecum in response to infections with non-typhoid Salmonella. Vet. Res. 45, 119 (2014).
Preusse, M., Tantawy, M. A., Klawonn, F., Schughart, K. & Pessler, F. Infection- and procedure-dependent effects on pulmonary gene expression in the early phase of influenza A virus infection in mice. BMC Microbiol. 13, 293 (2013).
Acknowledgements
We thank Dave Primm (Department of Surgery, University of Texas Southwestern Medical Center) for his critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Wu, R., Chen, F., Wang, N. et al. ACOD1 in immunometabolism and disease. Cell Mol Immunol 17, 822–833 (2020). https://doi.org/10.1038/s41423-020-0489-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-020-0489-5
Key words
This article is cited by
-
Immune-related transcriptomic and epigenetic reconfiguration in BV2 cells after lipopolysaccharide exposure: an in vitro omics integrative study
Inflammation Research (2024)
-
Immune Response Gene-1 [IRG1]/itaconate protect against multi-organ injury via inhibiting gasdermin D-mediated pyroptosis and inflammatory response
Inflammopharmacology (2024)
-
Microbiota promotes recruitment and pro-inflammatory response of caecal macrophages during E. tenella infection
Gut Pathogens (2023)
-
Sirt3 improves monosodium urate crystal-induced inflammation by suppressing Acod1 expression
Arthritis Research & Therapy (2023)
-
Novel long non-coding RNAs associated with inflammation and macrophage activation in human
Scientific Reports (2023)