Abstract
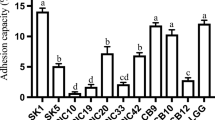
The objective of this study was to isolate and characterize lactic acid bacteria with probiotic potential in silages of different species of forage plants, cocoa beans, and artisanal salami. The obtained isolates were submitted to the following evaluations: (i) screening for tolerance to pH 2 and bile salts, (ii) genotypic identification of isolates, (iii) survival in simulated gastric and pancreatic conditions, (iv) antimicrobial activity, (v) antibiotic susceptibility and safety, and (vi) properties associated with adhesion capacity. A total of 82 isolates were obtained and were screened for pH 2.0 tolerance and capacity to growth in the presence of bile salts (1.0 and 2.0%). Only 19 strains simultaneously presented tolerance to pH 2.0 and bile salts. These 19 strains were evaluated for genetic profile by Box-PCR. Subsequently, the selected strains were subjected to partial sequencing of the 16S rRNA gene. The species Lactobacillus plantarum was prevalent. The identified strains were evaluated for survival under simulated gastric and pancreatic conditions. Some strains have shown tolerance in both conditions. Different strains showed variations in antimicrobial activity, susceptibility to antibiotics, and properties associated with adhesion (hydrophobicity, autoaggregation, coaggregation, and adhesion to CaCo2 cells). All strains were negative for hemolysis, DNase, gelatinase, and biogenic amine synthesis activity. The L. plantarum SBR64.7 strain can be considered the most promising for it presented the lowest viability reduction when exposed to gastric and pancreatic juices.


Similar content being viewed by others
References
Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, Calder PC, Sanders ME (2014) Expert consensus document. The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Ver Gastroenterol Hepatol 11:506–514. https://doi.org/10.1038/nrgastro.2017.75
Argyri AA, Zoumpopoulou G, Karatzas KA, Tsakalidou E, Nychas GJ, Panagou EZ, Tassou CC (2013) Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol 33(2):282–291. https://doi.org/10.1016/j.fm.2012.10.005
Floros G, Hatzikamari M, Litopoulou-Tzanetaki E, Tzanetakis N (2012) Probiotic and technological properties of facultatively heterofermentative lactobacilli from Greek traditional cheeses. Food Biotechnol 26(1):85–105. https://doi.org/10.1080/08905436.2011.645941
Santos TT, Ornellas RMS, Arcucio LB, Oliveira MM, Nicoli JR, Dias CV, Uetanabaro APT, Vinderola G (2016) Characterization of lactobacilli strains derived from cocoa fermentation in the south of Bahia for the development of probiotic cultures. LWT - Food Sci Technol 73:259–266. https://doi.org/10.1016/j.lwt.2016.06.003
Albuquerque TMR, Garcia EF, Araújo AO, Magnani M, Saarela M, Souza EL (2018) In vitro characterization of Lactobacillus strains isolated from fruit processing by-products as potential probiotics. Probiotics Antimicrob Proteins 10:704–716. https://doi.org/10.1007/s12602-017-9318-2
Shi Y, Cui X, Gu S, Yan X, Li R, Xia S, Chen H, Ge J (2019) Antioxidative and probiotic activities of lactic acid bacteria isolated from traditional artisanal milk cheese from Northeast China. Probiotic Antimicrob Proteins 11:1086–1099. https://doi.org/10.1007/s12602-018-9452-5
Sharma K, Attri S, Goel G (2019) Selection and evaluation of probiotic and functional characteristics of autochthonous lactic acid bacteria isolated from fermented wheat flour dough babroo. Probiotics Antimicrob Proteins 11:774–784. https://doi.org/10.1007/s12602-018-9466-z
Vasiee A, Behbahani BA, Yazdi FT, Mortazavi SA, Noorbakhsh H (2017) Diversity and probiotic potential of lactic acid bacteria isolated from horreh, a traditional Iranian fermented food. Probiotics Antimicrob Proteins 10:258–268. https://doi.org/10.1007/s12602-017-9282-x
Makete G, Aiyegoro OA, Thantsha S (2016) Isolation, identification and screening of potential probiotic bacteria in milk from south African Saanen goats. Probiotics Antimicrob Proteins 9(3):246–254. https://doi.org/10.1007/s12602-016-9247-5
Reda RM, Selim KM, El-Sayed M, El-Hady A (2018) In vitro selection and identification of potential probiotics isolated from the gastrointestinal tract of Nile Tilapia, Oreochromis niloticus. Probiotics Antimicrob Proteins 10:692–703. https://doi.org/10.1007/s12602-017-9314-6
Verso LL, Lessard M, Talbot G, Fernandez B, Fliss I (2018) Isolation and selection of potential probiotic bacteria from the pig gastrointestinal tract. Probiotics Antimicrob Proteins 10:299–312. https://doi.org/10.1007/s12602-017-9309-3
Pimm SL, Jenkins CN, Abell R, Brooks TM, Gittleman JL, Joppa LN, Raven PH, Roberts CM, Sexton JO (2014) The biodiversity of species and their rates of extinction, distribution, and protection. Science 344(6187):897–1000. https://doi.org/10.1126/science.1246752
Ramos CL, Thorsen L, Schwan RF, Jespersen L (2013) Strain-specific probiotics properties of Lactobacillus fermentum, Lactobacillus plantarum and Lactobacillus brevis isolates from Brazilian food products. Food Microbiol 36(1):22–29. https://doi.org/10.1016/j.fm.2013.03.010
Ramos CL, Sousa ESO, de Ribeiro J, Almeida TMM, Santos CCA, Abegg MA, Schwan RF (2015). Microbiological and chemical characteristics of tarubá, an indigenous beverage produced from solid cassava fermentation. Food Microbiol 49(0): 182–188. https://doi.org/10.1016/j.fm.2015.02.005
Puerari C, Magalhães-Guedes KT, Schwan RF (2015) Physicochemical and microbiological characterization of chicha, a rice-based fermented beverage produced by Umutina Brazilian Amerindians. Food Microbiol 46:210–217. https://doi.org/10.1016/j.fm.2014.08.009
Klocke M, Mundt K, Idler C, Mceniry J, O’kiely P, Barth S (2006) Monitoring Lactobacillus plantarum in grass silages with the aid of 16S rDNA-based quantitative real-time PCR assays. Syst Appl Microbiol 29:49–58. https://doi.org/10.1016/j.syapm.2005.06.001
Sandi S, Sari M L, Sahara E, Supriyadi A, Miksusanti Gofar N, Asmak (2019) Acid resistance test of probiotic isolated from silage forage swamp on in vitro digestive tract. Indones J Fundam App Chemi 15–20. https://doi.org/10.24845/ijfac.v4.i1.15
Soundharrajan I, Kim D, Kuppusamy P, Muthusamy K, Lee HJ, Choi KC (2019) Probiotic and Triticale silage fermentation potential of Pediococcus pentosaceus and Lactobacillus brevis and their impacts on pathogenic bacteria. Microorganisms 7:318. https://doi.org/10.3390/microorganisms7090318
Frederici CF, Campana R, Ciandrini E, Blasi G, Baffone W (2014) Identification and functional traits of lactic acid bacteria isolated from Ciauscolo salami produced in Central Italy. Meat Sci 98:575–584. https://doi.org/10.1016/j.meatsci.2014.05.019
Barbosa MS, Todorov SD, Ivanova I, Chobert J, Haert T, Franco BDGM (2015) Improving safety of salami by application of bacteriocins produced by an autochthonous Lactobacillus curvatus isolate. Food Microbiol 46:254–262. https://doi.org/10.1016/j.fm.2014.08.004
Schwan RF (1998) Cocoa fermentations conducted with a defined microbial cocktail inoculum. Appl Environ Microbiol 64(4):1477–1483. https://doi.org/10.1128/AEM.64.4.1477-1483.1998
Schwan RF, Wheals AE (2004) The microbiology of cocoa fermentation and its role in chocolate quality. Crit Revi Food Sci Nutr 44(4):205–221. https://doi.org/10.1080/10408690490464104
Oliveira JS, Costa K, Acurcio LB, Sandes SH, Cassali GD, Uetanabaro APT, Costa AM, Nicoli JR, Neumann E, Porto ALF (2018) In vitro and in vivo evaluation of two potential probiotic lactobacilli isolated from cocoa fermentation (Theobroma cacao L.). J Funct Foods 47:184–191. https://doi.org/10.1016/j.jff.2018.05.055
Koeuth T, Versalovic J, Lupski JR (1995) Differential subsequence conservation of interspersed repetitive Streptococcus pneumoniae BOX elements in diverse bacteria. Genome Res 5:408–418. https://doi.org/10.1101/gr.5.4.408
Heuer H, Krsek M, Baker P, Smalla K, Wellington EMH (1997) Analysis of actinomycete communities by specific amplification of genes encoding 16S RNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol 63:3233–3241
Lane DJ (1991) S/23S rRNA sequencing. In nucleic acid techniques in bacterial systematics (ed). Stackebrandt E, Goodfellow M, pp 115-175 New York: john Wiley & sons ltd.
Bautista-Gallego J, Arroyo-López FN, Rantsiou K, Jiménez-Díaz R (2013) Screening of lactic acid bacteria isolated from fermented table olives with probiotic potential. Food Res Int 50:135–142. https://doi.org/10.1016/j.foodres.2012.10.004
Ahn T, Kim GB, Lim KS, Baek YJ, Kim HU (2003) Desconjugation of bile salts by Lactobacillus acidophilus isolates. Int Dairy J 13(4):303–311. https://doi.org/10.1016/S0958-6946(02)00174-7
Marshall VM (1979) A note on screening hydrogen peroxide producing lactic acid bacteria using a non-toxic chromogen. J Appl Bacteriol 47:327–328. https://doi.org/10.1111/j.1365-2672.1979.tb01762.x
Tagg JR, Dajani AS, Wannamaker LW (1976) Bacteriocins of gram-positive bacteria. Bacteriol Rev 40:722–756
Hütt P, Shchepetova J, Lõivukene K, Kullisaar T, Mikelsaar M (2006) Antagonistic activity of probiotic lactobacilli and bifidobacteria against entero- and uropathogens. J Appl Microb 100(6):1324–1332. https://doi.org/10.1111/j.1365-2672.2006.02857.x
Tiago I, Teixeira I, Silva S, Chung P, Veríssimo A, Manaia CM (2004) Metabolic and genetic diversity of mesophilic and thermophilic bacteria isolated from composted municipal sludge on poly-e-caprolactones. Curr Microbiol 49:407–414. https://doi.org/10.1007/s00284-004-4353-0
Ben Omar N, Castro A, Lucas R, Abrioue H, Yousif NM, Franz CM, Holzapfel WH, Pérez-Pulido R, Martinez-Cañamero M, Gálves A (2004) Functional and safety aspects of enterococci isolated from different Spanish foods. Syst Appl Microbiol 27:118–130. https://doi.org/10.1078/0723-2020-00248
Semedo T, Santos MA, Martins P, Lopes MFS, Marques JJF, Tenreiro R, Crespo MTB (2003) Comparative study using type strains and clinical and food isolates to examine hemolytic activity and occurrence of the cyl operon in enterococci. J Clin Microbiol 41:2569–2576. https://doi.org/10.1128/JCM.41.6.2569-2576.2003
Yousif NMK, Dawyndt P, Abriouel H, Wijaya A, Schillinger U, Vancanneyt M, Swings J, Dirar HA, Holzapfel WH, Franz CMAP (2005) Molecular characterization, technological properties and safety aspects of enterococci from ‘Hussuwa’, an African fermented sorghum product. J Appl Microbiol 98:216–228. https://doi.org/10.1111/j.1365-2672.2004.02450.x
Divya JB, Varsha KK, Nampoothiri KM (2012) Newly isolated lactic acid bacteria with probiotic features for potential application in food industry. Appl Biochem Biotechnol 167:1314–1324. https://doi.org/10.1007/s12010-012-9561-7
Todorov SD, Furtado DN, Saad SMI, Tome E, Franco BDGM (2011) Potential beneficial properties of bacteriocin-producing lactic acid bacteria isolated from smoked salmon. J Appl Microbiol 110:971–986. https://doi.org/10.1111/j.1365-2672.2011.04950.x
Todorov SD, Botes M, Guigas C, Schillinger U, Wiid I, Wachsman MB, Holzapfel WH, Dicks LMT (2008) Boza, a natural source of probiotic lactic acid bacteria. J Appl Microbiol 104:465–477. https://doi.org/10.1111/j.1365-2672.2007.03558.x
Ranadheera CS, Evans CA, Adams MC, Baines SK (2014) Effect of dairy probiotic combinations on in vitro gastrointestinal tolerance, intestinal epithelial cell adhesion and cytokine secretion. J Funct Foods 8:18–25. https://doi.org/10.1016/j.jff.2014.02.022
De Angelis M, Gobbetti M (2004) Environmental stress responses in Lactobacillus: a review. Proteomics 4:106–122. https://doi.org/10.1002/pmic.200300497
Van De Gutche M, Serror P, Chervaux C, Smokvina T, Stanislav D (2002) Stress response in lactic acid bacteria. Anton Leeuw 82:187–216
Tokatli M, Gulgor G, Elmaci SB, Isleyen NA, Ozcelik F (2015) In vitro properties of potencial probiotic indigenous lactic acid bacteria originating from tradicional pickles. Biomed Res Int 2015:1–8. https://doi.org/10.1155/2015/315819
Gilliland SE, Staley TE, Bush LJ (1985) Importance of bile tolerance of Lactobacillus acidophilus used as a dietary adjunct. J Dairy Sci 67:3045–3051. https://doi.org/10.3168/jds.S0022-0302(84)81670-7
Leroy F, De Vuyst L (2004) Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci Technol 15:67–78. https://doi.org/10.1016/j.tifs.2003.09.004
Lee N-K, Paik H-D (2001) Partial characterization of lacticin NK24, a newly identified bacteriocin Lactococcus lactis NK24 isolated from Jeot-gal. Food Microbiol 18(1):17–24. https://doi.org/10.1006/fmic.2000.0368
Todorov SD, Dicks LMT (2007) Bacteriocin production by Lactobacillus pentosus ST712BZ isolated from Boza. Braz J Microbiol 38(1):166–172. https://doi.org/10.1590/S1517-83822007000100034
Azhar NS, Zin NHM, Haziyamin Hamid THTA (2017) Lactococcus Lactis strains A5 producing nisin-like bacteriocin active against gram positive and negative bacteria. Trop life Sci Res 28(2):107–118. https://doi.org/10.21315/tlsr2017.28.2.8
Rajoka MSR, Mehwish HM, Siddiq M, Haobin Z, Zhu J, Yan L, Shao D, Xu X, Shi J (2017) Identification, characterization, and probiotic potential of Lactobacillus rhamnosus isolated from human milk. LWT-Food Sci Technol 84:271–280. https://doi.org/10.1016/j.lwt.2017.05.055
Del Re B, Sgorbati B, Miglioli M, Palenzona D (2000) Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett Appl Microbiol 31(6):438–442. https://doi.org/10.1046/j.1365-2672.2000.00845.x
Vinderola CG, Reinheimer JA (2003) Lactic acid starter and probiotic bacteria: a comparative in vitro study of probiotic characteristics and biological barrier resistance. Food Res Int 36:895–904. https://doi.org/10.1016/S0963-9969(03)00098-X
De Vries MC, Vaughan EE, Kleerebezem M, De Vos WM (2006) Lactobacillus plantarum survival, functional and potential probiotic properties in the human intestinal tract. Int Dairy J 16:1018–1028. https://doi.org/10.1016/j.idairyj.2005.09.003
Bao Y, Zhang Y, Zhang Y, Liu Y, Wang S, Dong X, Wang Y, Zhang H (2010) Screening of potential probiotic properties of Lactobacillus fermentum isolated from traditional dairy products. Food Control 21:695–701. https://doi.org/10.1016/j.foodcont.2009.10.010
Iñiguez-Palomares C, Pérez-Morales R, Acedo-Félix (2008) Evaluation of probiotic properties in Lactobacillus isolated from small intestine of piglets. Rev Latinoam Microbiol 49:46–54
Pelletier C, Bouley C, Cayuela C, Bouttier S, Bourlioux P, Bellon-Fontaine MN (1997) Cell surface characteristics of Lactobacillus casei subsp. casei, Lactobacillus paracasei subsp paracasei, and Lactobacillus rhamnosus strains. Appl Environ Microbiol 63(5):1725–1731
Ehrmann MA, Kurzak P, Bauer J, Vogel RF (2002) Characterization of lactobacilli towards their use as probiotic adjuncts in poultry. J Appl Microbiol 92(5):966–975. https://doi.org/10.1046/j.1365-2672.2002.01608.x
Vera-Pingitore E, Jimenez ME, Dallagnol A, Belfiore C, Fontana C, Fontana P, Wright AV, Vignolo G, Plumed-Ferrer C (2016) Screening and characterization of potential probiotic and starter bacteria for plant fermentations. LWT-Food Sci Technol 71:288–294. https://doi.org/10.1016/j.lwt.2016.03.046
Sophatha B, Piwat S, Teanpaisan R (2020) Adhesion, anti-adhesion and aggregation properties relating to surface charges of selected Lactobacillus strains: study in Caco-2 and H357 cells. Arch Microbiol:1–9. https://doi.org/10.1007/s00203-020-01846-7
Olsen I, Progulske-Fox A (2015) Invasion of Porphyromonas gingivalis strains into vascular cells and tissue. J Oral Microbiol 7:28788. https://doi.org/10.3402/jom.v7.2878
Funding
This study was supported by Federal District Foundation for Research (project number—193.000.812 / 2015).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical Approval/Ethics Statement
The study was not conducted with animals and humans.
Conflict of Interest
The authors have declared no conflict of interest for this article.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
dos Santos Leandro, E., Ginani, V.C., de Alencar, E.R. et al. Isolation, Identification, and Screening of Lactic Acid Bacteria with Probiotic Potential in Silage of Different Species of Forage Plants, Cocoa Beans, and Artisanal Salami. Probiotics & Antimicro. Prot. 13, 173–186 (2021). https://doi.org/10.1007/s12602-020-09679-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-020-09679-y