Abstract
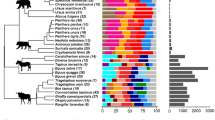
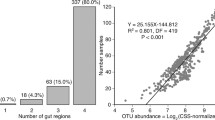
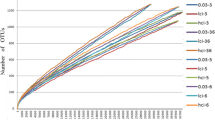
Most knowledge of the vertebrate gut microbiota comes from fecal samples; due to difficulties involved in sample collection, the upper intestinal microbiota is poorly understood in wild animals despite its potential to inform broad interpretations about host-gut microbe relationships under natural conditions. Here, we used 16S rRNA gene sequencing to characterize the microbiota of wild coyotes (Canis latrans) along the gastrointestinal tract, including samples from the duodenum, jejunum, ileum, caecum, ascending and descending colon, and feces. We used this intestinal profile to (1) quantify how intestinal site and individual identity interact to shape the microbiota in an uncontrolled setting, and (2) evaluate whether the fecal microbiota adequately represent other intestinal sites. Microbial communities in the large intestine were distinct from those in the small intestine, with higher diversity and a greater abundance of anaerobic taxa. Within each of the small and large intestine, individual identity explained significantly more among-sample variation than specific intestinal sites, revealing the importance of individual variation in the microbiota of free-living animals. Fecal samples were not an adequate proxy for studying upper intestinal environments, as they contained only half the amplicon sequence variants (ASVs) present in the small intestine at three- to four-fold higher abundances. Our study is a unique biogeographical investigation of the microbiota using free-living mammals rather than livestock or laboratory organisms and provides a foundational understanding of the gastrointestinal microbiota in a wild canid.





Similar content being viewed by others
Data Availability
The unfiltered sequence data used in this study has been deposited in the NCBI Short Read Archive under accession number PRJNA528765.
References
Lozupone C, Stomabaugh J, Gordon J et al (2012) Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. https://doi.org/10.1038/nature11550.Diversity
McFall-Ngai MJ, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ (2013) Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. 110:3229–3236. https://doi.org/10.1073/pnas.1218525110
Amato KR (2013) Co-evolution in context: the importance of studying gut microbiomes in wild animals. Microbiome Sci Med 1:10–29. https://doi.org/10.2478/micsm-2013-0002
Muegge BD, Kuczynski J, Knights D et al (2011) Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science (80- ) 332:970–974. https://doi.org/10.1126/science.1198719
Hicks AL, Lee KJ, Couto-Rodriguez M, Patel J, Sinha R, Guo C, Olson SH, Seimon A, Seimon TA, Ondzie AU, Karesh WB, Reed P, Cameron KN, Lipkin WI, Williams BL (2018) Gut microbiomes of wild great apes fluctuate seasonally in response to diet. Nat. Commun. 9:1786. https://doi.org/10.1038/s41467-018-04204-w
Sherrill-Mix S, McCormick K, Lauder A, Bailey A, Zimmerman L, Li Y, Django JBN, Bertolani P, Colin C, Hart JA, Hart TB, Georgiev AV, Sanz CM, Morgan DB, Atencia R, Cox D, Muller MN, Sommer V, Piel AK, Stewart FA, Speede S, Roman J, Wu G, Taylor J, Bohm R, Rose HM, Carlson J, Mjungu D, Schmidt P, Gaughan C, Bushman JI, Schmidt E, Bittinger K, Collman RG, Hahn BH, Bushman FD (2018) Allometry and ecology of the bilaterian gut microbiome. MBio 9:e00319–e00318. https://doi.org/10.1128/mBio.00319-18
Hanning I, Diaz-Sanchez S (2015) The functionality of the gastrointestinal microbiome in non-human animals. Microbiome 3:51. https://doi.org/10.1186/s40168-015-0113-6
Karasov WH, Douglas AE (2013) Comparative digestive physiology. Comp Physiol 3:741–783. https://doi.org/10.1002/cphy.c110054.Comparative
Hillman ET, Lu H, Yao T, Nakatsu CH (2017) Microbial ecology along the gastrointestinal tract. Microbes Environ. 32:300–313. https://doi.org/10.1264/jsme2.me17017
Sender R, Fuchs S, Milo R (2016) Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14:e1002533. https://doi.org/10.1371/journal.pbio.1002533
Pilla R, Suchodolski JS (2020) The role of the canine gut microbiome and metabolome in health and gastrointestinal disease. Front Vet Sci 6:498. https://doi.org/10.3389/fvets.2019.00498
Angelakis E, Armougom F, Carri F et al (2015) A metagenomic investigation of the duodenal microbiota reveals links with obesity. PLoS One 10:e0137784. https://doi.org/10.1371/journal.pone.0137784
Onishi JC, Campbell S, Moreau M, Patel F, Brooks AI, Zhou YX, Häggblom MM, Storch J (2017) Bacterial communities in the small intestine respond differently to those in the caecum and colon in mice fed low- and high-fat diets. Microbiology 163:1189–1197. https://doi.org/10.1099/mic.0.000496
Nagata N, Tohya M, Fukuda S, Suda W, Nishijima S, Takeuchi F, Ohsugi M, Tsujimoto T, Nakamura T, Shimomura A, Yanagisawa N, Hisada Y, Watanabe K, Imbe K, Akiyama J, Mizokami M, Miyoshi-Akiyama T, Uemura N, Hattori M (2019) Effects of bowel preparation on the human gut microbiome and metabolome. Sci. Rep. 9:4042. https://doi.org/10.1038/s41598-019-40182-9
Gu S, Chen D, Zhang J-N, Lv X, Wang K, Duan LP, Nie Y, Wu XL (2013) Bacterial community mapping of the mouse gastrointestinal tract. PLoS One 8:e74957. https://doi.org/10.1371/journal.pone.0074957
Han X, Shao H, Wang Y, Hu A, Chen R, Chen Q (2020) Composition of the bacterial community in the gastrointestinal tract of Kunming mice. Electron. J. Biotechnol. 43:16–22. https://doi.org/10.1016/j.ejbt.2019.11.003
Zhang L, Jiang X, Li A, Waqas M, Gao X, Li K, Xie G, Zhang J, Mehmood K, Zhao S, Wangdui B, Li J (2020) Characterization of the microbial community structure in intestinal segments of yak (Bos grunniens). Anaerobe 61:102115. https://doi.org/10.1016/j.anaerobe.2019.102115
Kim JH, Hong SW, Park BY, Yoo JG, Oh MH (2019) Characterisation of the bacterial community in the gastrointestinal tracts of elk (Cervus canadensis). Antonie van Leeuwenhoek, Int J Gen Mol Microbiol 112:225–235. https://doi.org/10.1007/s10482-018-1150-5
Thomas NA, Olvera-Ramírez AM, Abecia L, Adam CL, Edwards JE, Cox GF, Findlay PA, Destables E, Wood TA, McEwan NR (2019) Characterisation of the effect of day length, and associated differences in dietary intake, on the gut microbiota of Soay sheep. Arch. Microbiol. 201:889–896. https://doi.org/10.1007/s00203-019-01652-w
Willson NL, Van TTH, Lever J et al (2019) Characterisation of the intestinal microbiota of commercially farmed saltwater crocodiles, Crocodylus porosus. Appl. Microbiol. Biotechnol. 103:8977–8985. https://doi.org/10.1007/s00253-019-10143-3
Yasuda K, Oh K, Ren B, Tickle TL, Franzosa EA, Wachtman LM, Miller AD, Westmoreland SV, Mansfield KG, Vallender EJ, Miller GM, Rowlett JK, Gevers D, Huttenhower C, Morgan XC (2015) Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe 17:385–391. https://doi.org/10.1016/j.chom.2015.01.015
Liu Y, Zheng Z, Yu L, Wu S, Sun L, Wu S, Xu Q, Cai S, Qin N, Bao W (2019) Examination of the temporal and spatial dynamics of the gut microbiome in newborn piglets reveals distinct microbial communities in six intestinal segments. Sci. Rep. 9:3453. https://doi.org/10.1038/s41598-019-40235-z
Honneffer JB, Steiner JM, Lidbury JA, Suchodolski JS (2017) Variation of the microbiota and metabolome along the canine gastrointestinal tract. Metabolomics 13:26. https://doi.org/10.1007/s11306-017-1165-3
Yan W, Sun C, Zheng J, Wen C, Ji C, Zhang D, Chen Y, Hou Z, Yang N (2019) Efficacy of fecal sampling as a gut proxy in the study of chicken gut microbiota. Front. Microbiol. 10:2126. https://doi.org/10.3389/fmicb.2019.02126
Videvall E, Strandh M, Engelbrecht A, Cloete S, Cornwallis CK (2018) Measuring the gut microbiome in birds: comparison of faecal and cloacal sampling. Mol. Ecol. Resour. 18:424–434. https://doi.org/10.1111/1755-0998.12744
Ingala MR, Simmons NB, Wultsch C, Krampis K, Speer KA, Perkins SL (2018) Comparing microbiome sampling methods in a wild mammal: fecal and intestinal samples record different signals of host ecology, evolution. Front. Microbiol. 9:803. https://doi.org/10.3389/fmicb.2018.00803
Pascoe EL, Hauffe HC, Marchesi JR, Perkins SE (2017) Network analysis of gut microbiota literature: an overview of the research landscape in non-human animal studies. ISME J 11:2644–2651. https://doi.org/10.1038/ismej.2017.133
Wasimuddin MS, Melzheimer J et al (2017) Gut microbiomes of free-ranging and captive Namibian cheetahs: diversity, putative functions and occurrence of potential pathogens. Mol. Ecol. 26:5515–5527. https://doi.org/10.1111/mec.14278
Nelson TM, Rogers TL, Carlini AR, Brown MV (2013) Diet and phylogeny shape the gut microbiota of Antarctic seals: a comparison of wild and captive animals. Environ. Microbiol. 15:1132–1145. https://doi.org/10.1111/1462-2920.12022
Alessandri G, Milani C, Mancabelli L, Mangifesta M, Lugli GA, Viappiani A, Duranti S, Turroni F, Ossiprandi MC, van Sinderen D, Ventura M (2019) The impact of human-facilitated selection on the gut microbiota of domesticated mammals. FEMS Microbiol. Ecol. 95:1–13. https://doi.org/10.1093/femsec/fiz121
Laviad-Shitrit S, Izhaki I, Lalzar M, Halpern M (2019) Comparative analysis of intestine microbiota of four wild waterbird species. Front. Microbiol. 10:1911. https://doi.org/10.3389/fmicb.2019.01911
Suchodolski JS, Camacho J, Steiner JM (2008) Analysis of bacterial diversity in the canine duodenum, jejunum, ileum, and colon by comparative 16S rRNA gene analysis. FEMS Microbiol. Ecol. 66:567–578. https://doi.org/10.1111/j.1574-6941.2008.00521.x
Gehrt SD, Riley SPD (2010) Coyotes (Canis latrans). In: Gehrt SD, Riley SPD, Cypher BL (eds) Urban Carnivores: Ecology, Conflict, and Conservation. pp 79–95
Murray M, Edwards MA, Abercrombie B, St. Clair CC (2015) Poor health is associated with use of anthropogenic resources in an urban carnivore. Proc. R. Soc. B Biol. Sci. 282:20150009. https://doi.org/10.1098/rspb.2015.0009
Callahan BJ, Mcmurdie PJ, Rosen MJ et al (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
R Core Team (2019) R: a language and environment for statistical computing
Callahan BJ, McMurdie PJ, Holmes SP (2017) Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 11:2639–2643. https://doi.org/10.1038/ismej.2017.119
Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM (2014) Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42:633–642. https://doi.org/10.1093/nar/gkt1244
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. https://doi.org/10.1128/AEM.00062-07
Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ (2018) Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6:226. https://doi.org/10.1186/s40168-018-0605-2
Schliep KP (2011) phangorn: phylogenetic analysis in R. Bioinformatics 27:592–593. https://doi.org/10.1093/bioinformatics/btq706
Callahan BJ, Sankaran K, Fukuyama JA, et al (2016) Bioconductor workflow for microbiome data analysis: from raw reads to community analyses. F1000Research 5:1492. https://doi.org/10.12688/f1000research.8986.2
McMurdie PJ, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. https://doi.org/10.1371/journal.pone.0061217
Gloor GB, Macklaim JM, Pawlowsky-Glahn V, Egozcue JJ (2017) Microbiome datasets are compositional: and this is not optional. Front. Microbiol. 8:2224–2882. https://doi.org/10.1080/01904168209363016
Fernandes A, Macklaim JM, Linn T et al (2013) ANOVA-like differential expression (ALDEx) analysis for mixed population RNA-Seq. PLoS One 8:e67019. https://doi.org/10.1371/journal.pone.0067019
Bates D, Machler M, Bolker BM, Walker SC (2015) Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67. https://doi.org/10.18637/jss.v067.i01
Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C (2012) Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13:R79. https://doi.org/10.1186/gb-2012-13-9-r79
Oksanen J, Blanchet FG, Friendly M, et al (2018) Vegan: community ecology package
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235. https://doi.org/10.1128/AEM.71.12.8228
Liaw A, Wiener M (2002) Classification and regression by randomForest. R News 2:18–22
Barton K (2018) MuMIn: multi-model inference
Colston TJ, Jackson CR (2016) Microbiome evolution along divergent branches of the vertebrate tree of life: what is known and unknown. Mol. Ecol. 25:3776–3800. https://doi.org/10.1111/mec.13730
Zhang H, Chen L (2010) Phylogenetic analysis of 16S rRNA gene sequences reveals distal gut bacterial diversity in wild wolves (Canis lupus). Mol. Biol. Rep. 37:4013–4022. https://doi.org/10.1007/s11033-010-0060-z
Schmidt M, Unterer S, Suchodolski JS, Honneffer JB, Guard BC, Lidbury JA, Steiner JM, Fritz J, Kölle P (2018) The fecal microbiome and metabolome differs between dogs fed bones and raw food (BARF) diets and dogs fed commercial diets. PLoS One 13:e0201279. https://doi.org/10.1371/journal.pone.0201279
Deng P, Swanson KS (2015) Gut microbiota of humans, dogs and cats: current knowledge and future opportunities and challenges. Br. J. Nutr. 113:S6–S17. https://doi.org/10.1017/S0007114514002943
Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, Bultman SJ (2011) The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 13:517–526. https://doi.org/10.1016/j.cmet.2011.02.018
Mu C, Yang Y, Su Y, Zoetendal EG, Zhu W (2017) Differences in microbiota membership along the gastrointestinal tract of piglets and their differential alterations following an early-life antibiotic intervention. Front. Microbiol. 8:797. https://doi.org/10.3389/fmicb.2017.00797
Crespo-Piazuelo D, Estellé J, Revilla M, Criado-Mesas L, Ramayo-Caldas Y, Óvilo C, Fernández AI, Ballester M, Folch JM (2018) Characterization of bacterial microbiota compositions along the intestinal tract in pigs and their interactions and functions. Sci. Rep. 8:12727. https://doi.org/10.1038/s41598-018-30932-6
Wilkinson TJ, Cowan AA, Vallin HE, Onime LA, Oyama LB, Cameron SJ, Gonot C, Moorby JM, Waddams K, Theobald VJ, Leemans D, Bowra S, Nixey C, Huws SA (2017) Characterization of the microbiome along the gastrointestinal tract of growing turkeys. Front. Microbiol. 8:1089. https://doi.org/10.3389/fmicb.2017.01089
Amato KR, Yeoman CJ, Kent A, Righini N, Carbonero F, Estrada A, Rex Gaskins H, Stumpf RM, Yildirim S, Torralba M, Gillis M, Wilson BA, Nelson KE, White BA, Leigh SR (2013) Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J 7:1344–1353. https://doi.org/10.1038/ismej.2013.16
Park W (2018) Gut microbiomes and their metabolites shape human and animal health. J. Microbiol. 56:151–153. https://doi.org/10.1007/s12275-018-0577-8
Rosenfeld CS (2017) Gut dysbiosis in animals due to environmental chemical exposures. Front. Cell. Infect. Microbiol. 7:396. https://doi.org/10.3389/fcimb.2017.00396
Wong K, Shaw TI, Oladeinde A, Glenn TC, Oakley B, Molina M (2016) Rapid microbiome changes in freshly deposited cow feces under field conditions. Front. Microbiol. 7:500. https://doi.org/10.3389/fmicb.2016.00500
Dressman JB (1986) Comparison of canine and human gastrointestinal physiology. Pharm. Res. 3:123–131. https://doi.org/10.1023/A:1016353705970
Gaulke CA, Arnold HK, Humphreys IR, Kembel SW, O’Dwyer JP, Sharpton TJ (2018) Ecophylogenetics clarifies the evolutionary association between mammals and their gut microbiota. MBio 9:e01348–e01318. https://doi.org/10.1128/mBio.01348-18
Robinson JWL, Menge H, Sepúlveda FV, Mirkovitch V (1977) Functional and structural characteristics of the jejunum and ileum in the dog and the rat. Digestion 15:188–199. https://doi.org/10.1159/000198003
Laganiere S, Berteloot A, Maestracci D (1984) Digestive and absorptive functions along the dog small intestine: comparative distributions in relation to biochemical and morphological parameters. Comp. Biochem. Physiol. 79A:463–472. https://doi.org/10.1016/0300-9629(84)90547-4
Murray M, Cembrowski A, Latham ADM, Lukasik VM, Pruss S, St Clair CC (2015) Greater consumption of protein-poor anthropogenic food by urban relative to rural coyotes increases diet breadth and potential for human-wildlife conflict. Ecography (Cop) 38:001–008. https://doi.org/10.1111/ecog.01128
Metzler-Zebeli BU, Schmitz-Esser S, Mann E, Grüll D, Molnar T, Zebeli Q (2015) Adaptation of the cecal bacterial microbiome of growing pigs in response to resistant starch type 4. Appl. Environ. Microbiol. 81:8489–8499. https://doi.org/10.1128/aem.02756-15
Zhu Y, Wang C, Li F (2015) Impact of dietary fiber/starch ratio in shaping caecal microbiota in rabbits. Can. J. Microbiol. 61:771–784. https://doi.org/10.1139/cjm-2015-0201
Tao S, Tian P, Luo Y, Tian J, Hua C, Geng Y, Cong R, Ni Y, Zhao R (2017) Microbiome-metabolome responses to a high-grain diet associated with the hind-gut health of goats. Front. Microbiol. 8:1764. https://doi.org/10.3389/fmicb.2017.01764
Galley JD, Yu Z, Kumar P, Dowd SE, Lyte M, Bailey MT (2015) The structures of the colonic mucosa-associated and luminal microbial communities are distinct and differentially affected by a prolonged murine stressor. Gut Microbes 5:748–760. https://doi.org/10.4161/19490976.2014.972241
Ringel Y, Maharshak N, Ringel-Kulka T, Wolber EA, Sartor RB, Carroll IM (2015) High throughput sequencing reveals distinct microbial populations within the mucosal and luminal niches in healthy individuals. Gut Microbes 6:173–181. https://doi.org/10.1080/19490976.2015.1044711
Yeoman CJ, Ishaq SL, Bichi E, Olivo SK, Lowe J, Aldridge BM (2018) Biogeographical differences in the influence of maternal microbial sources on the early successional development of the bovine neonatal gastrointestinal tract. Sci. Rep. 8:3197. https://doi.org/10.1038/s41598-018-21440-8
Acknowledgments
Coyote carcasses were provided by Bill, Duncan, and Malcolm Abercrombie of Animal Damage Control, Inc., and SS would like to thank Dana Sanderson and several undergraduate volunteers at the University of Alberta for assistance with coyote necropsies.
Code Availability
The R scripts and all material(code and workspace) is available (present tense) in the linked GitHub repository https://github.com/sasugden/Coyote_intestinal_biogeography.
Funding
This study was supported by Discovery Grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) to CCSC (RGPIN-2017-05915) and LYS (RGPIN-2019-04399).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
All three authors have agreed to submit this work for publication in Microbial Ecology.
Electronic Supplementary Material
ESM 1
(DOCX 3104 kb)
Rights and permissions
About this article
Cite this article
Sugden, S., St. Clair, C.C. & Stein, L.Y. Individual and Site-Specific Variation in a Biogeographical Profile of the Coyote Gastrointestinal Microbiota. Microb Ecol 81, 240–252 (2021). https://doi.org/10.1007/s00248-020-01547-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-020-01547-0