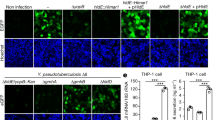
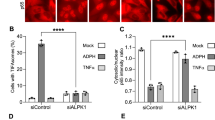
Abstract
The innate immune response constitutes the first line of defense against pathogens. It involves the recognition of pathogen-associated molecular patterns (PAMPs) by pathogen recognition receptors (PRRs), the production of inflammatory cytokines and the recruitment of immune cells to infection sites. Recently, ADP-heptose, a soluble intermediate of the lipopolysaccharide biosynthetic pathway in Gram-negative bacteria, has been identified by several research groups as a PAMP. Here, we recapitulate the evidence that led to this identification and discuss the controversy over the immunogenic properties of heptose 1,7-bisphosphate (HBP), another bacterial heptose previously defined as an activator of innate immunity. Then, we describe the mechanism of ADP-heptose sensing by alpha-protein kinase 1 (ALPK1) and its downstream signaling pathway that involves the proteins TIFA and TRAF6 and induces the activation of NF-κB and the secretion of inflammatory cytokines. Finally, we discuss possible delivery mechanisms of ADP-heptose in cells during infection, and propose new lines of thinking to further explore the roles of the ADP-heptose/ALPK1/TIFA axis in infections and its potential implication in the control of intestinal homeostasis.



Similar content being viewed by others
Abbreviations
- ADP-heptose:
-
ADP-L-glycero-β-D-manno-heptose
- ADP-heptose 7P:
-
ADP-heptose 7-phosphate
- AIDA:
-
Adhesin involved in diffuse adherence
- AKT:
-
AK strain-transforming protein/protein kinase B
- ALPK1:
-
Alpha-protein kinase 1
- CD:
-
Cluster of differentiation
- CLR:
-
C-type lectin receptor
- dsRNA:
-
Double-stranded RNA
- FHA:
-
Forkhead-associated domain
- GM-CSF:
-
Granulocyte–macrophage colony stimulating factor
- HBP:
-
D-glycero-β-D-manno-heptose 1,7-bisphosphate
- HEK:
-
Human embryonic kidney
- HIV:
-
Human immunodeficiency virus
- HMP1:
-
D-glycero-β-D-manno-heptose 1-monophosphate
- IFN:
-
Interferon
- IKK:
-
IκB kinase
- IL:
-
Interleukin
- IP:
-
Interferon-γ induced protein
- IκB:
-
Inhibitor of κB
- Kdo:
-
3-Deoxy-D-manno-oct-2-ulosonic acid
- KIF:
-
Kinesin superfamily protein
- LPS:
-
Lipopolysaccharide
- MCP:
-
Monocyte chemoattractant protein
- MIP:
-
Macrophage inflammatory protein
- MS:
-
Mass spectrometry
- NEMO:
-
NF-κB essential modulator
- NF-κB:
-
Nuclear-factor kappa B
- NLR:
-
NOD-like receptor
- NLRP3:
-
NOD-, LRR- and pyrin domain-containing protein 3
- NMNAT:
-
Nicotinamide mononucleotide adenylyltransferase
- NOD:
-
Nucleotide-binding and oligomerization domain
- NTD:
-
N-terminal domain
- PAMP:
-
Pathogen-associated molecular pattern
- PRR:
-
Pathogen recognition receptor
- pT9:
-
Phospho-threonine 9
- RANTES:
-
Regulated on activation, normal T-cell expressed and secreted
- RIG:
-
Retinoid acid-inducible gene
- RLK:
-
Receptor-like kinase
- RLR:
-
RIG-like receptor
- T3/4SS:
-
Type 3/4 secretion system
- T9:
-
Threonine 9
- TAB:
-
TAK-1-binding protein
- TAK:
-
Transforming growth factor β-activated kinase 1
- Th1:
-
Type 1 helper T cell
- Th17:
-
Type 17 helper T cell
- TIFA:
-
TRAF-interacting protein with FHA domain-containing protein A
- TLR:
-
Toll-like receptor
- TNFα:
-
Tumor necrosis factor alpha
- TRAF:
-
Tumor necrosis factor receptor-associated factor
- Treg:
-
Regulatory T cell
- TRIM:
-
Tripartite motif
- Wt:
-
Wild type
- ZCCHC:
-
Zinc finger CCHC-type containing protein
References
Creagh EM, O’Neill LAJ (2006) TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol 27:352–357. https://doi.org/10.1016/j.it.2006.06.003
Akira S, Uematsu S, Takeuchi O (2006) Pathogen recognition and innate immunity. Cell 124:783–801. https://doi.org/10.1016/j.cell.2006.02.015
Baccala R, Gonzalez-Quintial R, Lawson BR et al (2009) Sensors of the innate immune system: their mode of action. Nat Rev Rheumatol 5:448–456. https://doi.org/10.1038/nrrheum.2009.136
Muñoz-Wolf N, Lavelle EC (2016) Innate immune receptors. Methods Mol Biol Clifton NJ 1417:1–43. https://doi.org/10.1007/978-1-4939-3566-6_1
Poltorak A, He X, Smirnova I et al (1998) Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282:2085–2088. https://doi.org/10.1126/science.282.5396.2085
Gewirtz AT, Navas TA, Lyons S et al (1950) (2001) Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol Baltim Md 167:1882–1885. https://doi.org/10.4049/jimmunol.167.4.1882
Brightbill HD, Libraty DH, Krutzik SR et al (1999) Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science 285:732–736. https://doi.org/10.1126/science.285.5428.732
Hirschfeld M, Kirschning CJ, Schwandner R et al (1950) (1999) Cutting edge: inflammatory signaling by Borrelia burgdorferi lipoproteins is mediated by toll-like receptor 2. J Immunol Baltim Md 163:2382–2386
Bauer S, Kirschning CJ, Häcker H et al (2001) Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc Natl Acad Sci U S A 98:9237–9242. https://doi.org/10.1073/pnas.161293498
Ishii KJ, Coban C, Kato H et al (2006) A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol 7:40–48. https://doi.org/10.1038/ni1282
Chamaillard M, Hashimoto M, Horie Y et al (2003) An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol 4:702–707. https://doi.org/10.1038/ni945
Girardin SE, Boneca IG, Carneiro LAM et al (2003) Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science 300:1584–1587. https://doi.org/10.1126/science.1084677
Girardin SE, Boneca IG, Viala J et al (2003) Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem 278:8869–8872. https://doi.org/10.1074/jbc.C200651200
Inohara N, Ogura Y, Fontalba A et al (2003) Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn’s disease. J Biol Chem 278:5509–5512. https://doi.org/10.1074/jbc.C200673200
Yoneyama M, Kikuchi M, Natsukawa T et al (2004) The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5:730–737. https://doi.org/10.1038/ni1087
Sato K, Yang X, Yudate T et al (2006) Dectin-2 is a pattern recognition receptor for fungi that couples with the Fc receptor gamma chain to induce innate immune responses. J Biol Chem 281:38854–38866. https://doi.org/10.1074/jbc.M606542200
Wells CA, Salvage-Jones JA, Li X et al (1950) (2008) The macrophage-inducible C-type lectin, mincle, is an essential component of the innate immune response to Candida albicans. J Immunol Baltim Md 180:7404–7413. https://doi.org/10.4049/jimmunol.180.11.7404
Janeway CA, Medzhitov R (2002) Innate immune recognition. Annu Rev Immunol 20:197–216. https://doi.org/10.1146/annurev.immunol.20.083001.084359
García-Weber D, Dangeard AS, Cornil J, et al (2018) ADP-heptose is a newly identified pathogen-associated molecular pattern of Shigella flexneri. EMBO Rep https://doi.org/10.15252/embr.201846943
Zhou P, She Y, Dong N et al (2018) Alpha-kinase 1 is a cytosolic innate immune receptor for bacterial ADP-heptose. Nature 561:122–126. https://doi.org/10.1038/s41586-018-0433-3
Pfannkuch L, Hurwitz R, Traulsen J et al (2019) ADP heptose, a novel pathogen-associated molecular pattern identified in Helicobacter pylori. FASEB J Off Publ Fed Am Soc Exp Biol. https://doi.org/10.1096/fj.201802555R
Raetz CRH (1990) Biochemistry of endotoxins. Annu Rev Biochem 59:129–170. https://doi.org/10.1146/annurev.bi.59.070190.001021
Shi J, Zhao Y, Wang Y et al (2014) Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514:187–192. https://doi.org/10.1038/nature13683
Park BS, Lee J-O (2013) Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med 45:e66. https://doi.org/10.1038/emm.2013.97
Kunin CM, Beard MV (1963) Serological studies of o antigens of Escherichia Coli by means of the hemagglutination test. J Bacteriol 85:541–548
Endotoxin in Health and Disease. In: CRC Press. https://www.routledge.com/Endotoxin-in-Health-and-Disease/Brade/p/book/9780824719449. Accessed 11 June 2020
Møller AK, Leatham MP, Conway T et al (2003) An Escherichia coli MG1655 lipopolysaccharide deep-rough core mutant grows and survives in mouse cecal mucus but fails to colonize the mouse large intestine. Infect Immun 71:2142–2152. https://doi.org/10.1128/iai.71.4.2142-2152.2003
Xu D, Zhang W, Zhang B et al (2016) Characterization of a biofilm-forming Shigella flexneri phenotype due to deficiency in Hep biosynthesis. PeerJ 4:e2178. https://doi.org/10.7717/peerj.2178
Martinić M, Hoare A, Contreras I, Álvarez SA (2011) Contribution of the lipopolysaccharide to resistance of Shigella flexneri 2a to extreme acidity. PLoS ONE. https://doi.org/10.1371/journal.pone.0025557
Coleman WG, Deshpande KS (1985) New cysE-pyrE-linked rfa mutation in Escherichia coli K-12 that results in a heptoseless lipopolysaccharide. J Bacteriol 161:1209–1214
Valvano MA, Marolda CL, Bittner M et al (2000) The rfaE gene from Escherichia coli encodes a bifunctional protein involved in biosynthesis of the lipopolysaccharide core precursor ADP-L-glycero-d-manno-heptose. J Bacteriol 182:488–497. https://doi.org/10.1128/jb.182.2.488-497.2000
Gronow S, Brade H (2001) Lipopolysaccharide biosynthesis: which steps do bacteria need to survive? J Endotoxin Res 7:3–23
Shih GC, Kahler CM, Carlson RW et al (2001) gmhX, a novel gene required for the incorporation of L-glycero-D-manno-heptose into lipooligosaccharide in Neisseria meningitidis. Microbiol Read Engl 147:2367–2377. https://doi.org/10.1099/00221287-147-8-2367
Kneidinger B, Marolda C, Graninger M et al (2002) Biosynthesis pathway of ADP-L-glycero-beta-D-manno-heptose in Escherichia coli. J Bacteriol 184:363–369. https://doi.org/10.1128/jb.184.2.363-369.2002
Valvano MA, Messner P, Kosma P (2002) Novel pathways for biosynthesis of nucleotide-activated glycero-manno-heptose precursors of bacterial glycoproteins and cell surface polysaccharides. Microbiol Read Engl 148:1979–1989. https://doi.org/10.1099/00221287-148-7-1979
Kim C-H (2003) A Salmonella typhimurium rfaE mutant recovers invasiveness for human epithelial cells when complemented by wild type rfaE (controlling biosynthesis of ADP-L-glycero-D-mannoheptose-containing lipopolysaccharide). Mol Cells 15:226–232
McArthur F, Andersson CE, Loutet S et al (2005) Functional analysis of the glycero-manno-heptose 7-phosphate kinase domain from the bifunctional HldE protein, which is involved in ADP-L-glycero-D-manno-heptose biosynthesis. J Bacteriol 187:5292–5300. https://doi.org/10.1128/JB.187.15.5292-5300.2005
Malott RJ, Keller BO, Gaudet RG et al (2013) Neisseria gonorrhoeae-derived heptose elicits an innate immune response and drives HIV-1 expression. Proc Natl Acad Sci U S A 110:10234–10239. https://doi.org/10.1073/pnas.1303738110
Pokorny B, Kosma P (2016) Synthesis of 5-O-oligoglucosyl extended α-(2→4)-Kdo disaccharides corresponding to inner core fragments of Moraxellaceae lipopolysaccharides. Carbohydr Res 422:5–12. https://doi.org/10.1016/j.carres.2015.12.009
Kadrmas JL, Brozek KA, Raetz CR (1996) Lipopolysaccharide core glycosylation in Rhizobium leguminosarum. An unusual mannosyl transferase resembling the heptosyl transferase I of Escherichia coli. J Biol Chem 271:32119–32125
Kay W, Petersen BO, Duus JØ et al (2006) Characterization of the lipopolysaccharide and beta-glucan of the fish pathogen Francisella victoria. FEBS J 273:3002–3013. https://doi.org/10.1111/j.1742-4658.2006.05311.x
Knirel YA, Moll H, Zähringer U (1996) Structural study of a highly O-acetylated core of Legionella pneumophila serogroup 1 lipopolysaccharide. Carbohydr Res 293:223–234. https://doi.org/10.1016/0008-6215(96)00194-2
Moreno E, Pitt MW, Jones LM et al (1979) Purification and characterization of smooth and rough lipopolysaccharides from Brucella abortus. J Bacteriol 138:361–369
Okan NA, Kasper DL (2013) The atypical lipopolysaccharide of Francisella. Carbohydr Res 378:79–83. https://doi.org/10.1016/j.carres.2013.06.015
de Vries SPW, Bootsma HJ, Hays JP, Hermans PWM (2009) Molecular aspects of Moraxella catarrhalis pathogenesis. Microbiol Mol Biol Rev MMBR 73:389–406. https://doi.org/10.1128/MMBR.00007-09
Hajjar AM, Harvey MD, Shaffer SA et al (2006) Lack of in vitro and in vivo recognition of Francisella tularensis subspecies lipopolysaccharide by Toll-like receptors. Infect Immun 74:6730–6738. https://doi.org/10.1128/IAI.00934-06
Tang W, Guo Z, Cao Z et al (2018) d-Sedoheptulose-7-phosphate is a common precursor for the heptoses of septacidin and hygromycin B. Proc Natl Acad Sci U S A 115:2818–2823. https://doi.org/10.1073/pnas.1711665115
Gaudet RG, Sintsova A, Buckwalter CM et al (2015) Innate immunity. Cytosolic detection of the bacterial metabolite HBP activates TIFA-dependent innate immunity. Science 348:1251–1255. https://doi.org/10.1126/science.aaa4921
Milivojevic M, Dangeard A-S, Kasper CA et al (2017) ALPK1 controls TIFA/TRAF6-dependent innate immunity against heptose-1,7-bisphosphate of gram-negative bacteria. PLoS Pathog 13:e1006224. https://doi.org/10.1371/journal.ppat.1006224
Stein SC, Faber E, Bats SH et al (2017) Helicobacter pylori modulates host cell responses by CagT4SS-dependent translocation of an intermediate metabolite of LPS inner core heptose biosynthesis. PLoS Pathog 13:e1006514. https://doi.org/10.1371/journal.ppat.1006514
Gall A, Gaudet RG, Gray-Owen SD, Salama NR (2017) TIFA Signaling in Gastric Epithelial Cells Initiates the cag Type 4 Secretion System-Dependent Innate Immune Response to Helicobacter pylori Infection. mBio https://doi.org/10.1128/mBio.01168-17
Zimmermann S, Pfannkuch L, Al-Zeer MA et al (2017) ALPK1- and TIFA-dependent innate immune response triggered by the Helicobacter pylori Type IV Secretion System. Cell Rep 20:2384–2395. https://doi.org/10.1016/j.celrep.2017.08.039
Adekoya IA, Guo CX, Gray-Owen SD et al (1950) (2018) d-Glycero-β-d-Manno-heptose 1-phosphate and d-glycero-β-d-manno-heptose 1,7-biphosphate are both innate immune agonists. J Immunol Baltim Md 201:2385–2391. https://doi.org/10.4049/jimmunol.1801012
Kanamori M, Suzuki H, Saito R et al (2002) T2BP, a novel TRAF2 binding protein, can activate NF-kappaB and AP-1 without TNF stimulation. Biochem Biophys Res Commun 290:1108–1113. https://doi.org/10.1006/bbrc.2001.6315
Takatsuna H, Kato H, Gohda J et al (2003) Identification of TIFA as an adapter protein that links tumor necrosis factor receptor-associated factor 6 (TRAF6) to interleukin-1 (IL-1) receptor-associated kinase-1 (IRAK-1) in IL-1 receptor signaling. J Biol Chem 278:12144–12150. https://doi.org/10.1074/jbc.M300720200
Huang C-CF, Weng J-H, Wei T-YW et al (2012) Intermolecular binding between TIFA-FHA and TIFA-pT mediates tumor necrosis factor alpha stimulation and NF-κB activation. Mol Cell Biol 32:2664–2673. https://doi.org/10.1128/MCB.00438-12
Weng J-H, Hsieh Y-C, Huang C-CF et al (2015) Uncovering the mechanism of forkhead-associated domain-mediated TIFA oligomerization that plays a central role in immune responses. Biochemistry 54:6219–6229. https://doi.org/10.1021/acs.biochem.5b00500
Ea C-K, Sun L, Inoue J-I, Chen ZJ (2004) TIFA activates IkappaB kinase (IKK) by promoting oligomerization and ubiquitination of TRAF6. Proc Natl Acad Sci U S A 101:15318–15323. https://doi.org/10.1073/pnas.0404132101
Gaudet RG, Guo CX, Molinaro R et al (2017) Innate recognition of intracellular bacterial growth is driven by the TIFA-dependent cytosolic surveillance pathway. Cell Rep 19:1418–1430. https://doi.org/10.1016/j.celrep.2017.04.063
Carson D, Barry R, Hopkins EGD et al (2020) Citrobacter rodentium induces rapid and unique metabolic and inflammatory responses in mice suffering from severe disease. Cell Microbiol 22:e13126. https://doi.org/10.1111/cmi.13126
Hillier LW, Graves TA, Fulton RS et al (2005) Generation and annotation of the DNA sequences of human chromosomes 2 and 4. Nature 434:724–731. https://doi.org/10.1038/nature03466
Scheeff ED, Bourne PE (2005) Structural evolution of the protein kinase-like superfamily. PLoS Comput Biol 1:e49. https://doi.org/10.1371/journal.pcbi.0010049
Zipfel C (2014) Plant pattern-recognition receptors. Trends Immunol 35:345–351. https://doi.org/10.1016/j.it.2014.05.004
Greeff C, Roux M, Mundy J, Petersen M (2012) Receptor-like kinase complexes in plant innate immunity. Front Plant Sci 3:209. https://doi.org/10.3389/fpls.2012.00209
Dardick C, Ronald P (2006) Plant and animal pathogen recognition receptors signal through non-RD kinases. PLoS Pathog 2:e2. https://doi.org/10.1371/journal.ppat.0020002
Chattopadhyay S, Sen GC (2014) Tyrosine phosphorylation in toll-like receptor signaling. Cytokine Growth Factor Rev 25:533–541. https://doi.org/10.1016/j.cytogfr.2014.06.002
Samuel CE (1993) The eIF-2 alpha protein kinases, regulators of translation in eukaryotes from yeasts to humans. J Biol Chem 268:7603–7606
Balachandran S, Roberts PC, Brown LE et al (2000) Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13:129–141. https://doi.org/10.1016/s1074-7613(00)00014-5
Dey M, Mann BR, Anshu A, Mannan MA (2014) Activation of protein kinase PKR requires dimerization-induced cis-phosphorylation within the activation loop. J Biol Chem 289:5747–5757. https://doi.org/10.1074/jbc.M113.527796
Middelbeek J, Clark K, Venselaar H et al (2010) The alpha-kinase family: an exceptional branch on the protein kinase tree. Cell Mol Life Sci CMLS 67:875–890. https://doi.org/10.1007/s00018-009-0215-z
Lin T-Y, Wei T-YW, Li S et al (2016) TIFA as a crucial mediator for NLRP3 inflammasome. Proc Natl Acad Sci U S A 113:15078–15083. https://doi.org/10.1073/pnas.1618773114
Wei T-YW, Wu P-Y, Wu T-J et al (2017) Aurora A and NF-κB survival pathway drive chemoresistance in acute myeloid leukemia via the TRAF-interacting protein TIFA. Cancer Res 77:494–508. https://doi.org/10.1158/0008-5472.CAN-16-1004
Oeckinghaus A, Hayden MS, Ghosh S (2011) Crosstalk in NF-κB signaling pathways. Nat Immunol 12:695–708. https://doi.org/10.1038/ni.2065
Lee C-P, Chiang S-L, Ko AM-S et al (2016) ALPK1 phosphorylates myosin IIA modulating TNF-α trafficking in gout flares. Sci Rep 6:25740. https://doi.org/10.1038/srep25740
Matsumura T, Semba K, Azuma S et al (2004) TIFAB inhibits TIFA, TRAF-interacting protein with a forkhead-associated domain. Biochem Biophys Res Commun 317:230–234. https://doi.org/10.1016/j.bbrc.2004.03.030
Matsumura T, Kawamura-Tsuzuku J, Yamamoto T et al (2009) TRAF-interacting protein with a forkhead-associated domain B (TIFAB) is a negative regulator of the TRAF6-induced cellular functions. J Biochem (Tokyo) 146:375–381. https://doi.org/10.1093/jb/mvp080
Minoda Y, Saeki K, Aki D et al (2006) A novel Zinc finger protein, ZCCHC11, interacts with TIFA and modulates TLR signaling. Biochem Biophys Res Commun 344:1023–1030. https://doi.org/10.1016/j.bbrc.2006.04.006
Huang W-C, Liao J-H, Hsiao T-C et al (2019) Binding and enhanced binding between key immunity proteins TRAF6 and TIFA. Chembiochem Eur J Chem Biol 20:140–146. https://doi.org/10.1002/cbic.201800436
Yin Q, Lin S-C, Lamothe B et al (2009) E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol 16:658–666. https://doi.org/10.1038/nsmb.1605
Parsot C (2009) Shigella type III secretion effectors: how, where, when, for what purposes? Curr Opin Microbiol 12:110–116. https://doi.org/10.1016/j.mib.2008.12.002
Kuehl CJ, Dragoi A-M, Talman A, Agaisse H (2015) Bacterial spread from cell to cell: beyond actin-based motility. Trends Microbiol 23:558–566. https://doi.org/10.1016/j.tim.2015.04.010
Kasper CA, Sorg I, Schmutz C et al (2010) Cell-cell propagation of NF-κB transcription factor and MAP kinase activation amplifies innate immunity against bacterial infection. Immunity 33:804–816. https://doi.org/10.1016/j.immuni.2010.10.015
Kawai T, Akira S (2007) Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med 13:460–469. https://doi.org/10.1016/j.molmed.2007.09.002
Platnich JM, Muruve DA (2019) NOD-like receptors and inflammasomes: A review of their canonical and non-canonical signaling pathways. Arch Biochem Biophys 670:4–14. https://doi.org/10.1016/j.abb.2019.02.008
Bleich A, Büchler G, Beckwith J et al (2010) Cdcs1 a major colitis susceptibility locus in mice; subcongenic analysis reveals genetic complexity. Inflamm Bowel Dis 16:765–775. https://doi.org/10.1002/ibd.21146
Boulard O, Kirchberger S, Royston DJ et al (2012) Identification of a genetic locus controlling bacteria-driven colitis and associated cancer through effects on innate inflammation. J Exp Med 209:1309–1324. https://doi.org/10.1084/jem.20120239
Ryzhakov G, West NR, Franchini F et al (2018) Alpha kinase 1 controls intestinal inflammation by suppressing the IL-12/Th1 axis. Nat Commun 9:3797. https://doi.org/10.1038/s41467-018-06085-5
Caruso R, Warner N, Inohara N, Núñez G (2014) NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity 41:898–908. https://doi.org/10.1016/j.immuni.2014.12.010
Kuo T-M, Hsu H-T, Chung C-M et al (2016) Enhanced alpha-kinase 1 accelerates multiple early nephropathies in streptozotocin-induced hyperglycemic mice. Biochim Biophys Acta 1862:2034–2042. https://doi.org/10.1016/j.bbadis.2016.08.010
Strietz J, Stepputtis SS, Preca B-T, et al (2016) ERN1 and ALPK1 inhibit differentiation of bi-potential tumor-initiating cells in human breast cancer. Oncotarget 7:83278–83293. https://doi.org/10.18632/oncotarget.13086
Li C, Kuang L, Zhu B et al (2017) Identification of prognostic risk factors of acute lymphoblastic leukemia based on mRNA expression profiling. Neoplasma 64:494–501. https://doi.org/10.4149/neo_2017_402
Ji C, Lin S, Yao D et al (2019) Identification of promising prognostic genes for relapsed acute lymphoblastic leukemia. Blood Cells Mol Dis 77:113–119. https://doi.org/10.1016/j.bcmd.2019.04.010
Chen P-K, Hua C-H, Hsu H-T et al (2019) ALPK1 expression is associated with lymph node metastasis and tumor growth in oral squamous cell carcinoma patients. Am J Pathol 189:190–199. https://doi.org/10.1016/j.ajpath.2018.09.003
Acknowledgements
We thank Anne-Sophie Dangeard and Veronica Teixeira for critically reviewing the manuscript.
Funding
We gratefully acknowledge financial support from the Agence Nationale de la Recherche (Grants no: ANR‐14‐ACHN‐0029‐01 and ANR‐17‐CE15‐0006, including postdoctoral fellowships to DGW) and from Fondation ARC pour la Recherche sur le Cancer (Grant No: ARC—PJA20171206187).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
García-Weber, D., Arrieumerlou, C. ADP-heptose: a bacterial PAMP detected by the host sensor ALPK1. Cell. Mol. Life Sci. 78, 17–29 (2021). https://doi.org/10.1007/s00018-020-03577-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-020-03577-w