Abstract
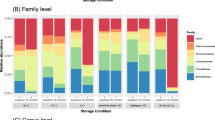
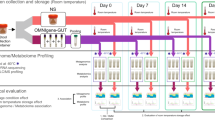
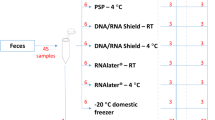
The gut microbiome, which is symbiotic within the human body, assists in human digestion. It plays significant roles in identifying intestinal disease as well as in maintaining a healthy body with functional immune and metabolic activities. To confirm the consistency of fecal intestinal microbial research, it is necessary to study the changes in intestinal microbial flora according to the fecal collection solution and storage period. We collected fecal samples from three healthy Korean adults. To examine the efficacy of fecal collection solution, we used NBgene-Gut, OMNIgene-Gut, 70% ethanol (Ethanol-70%), and RNAlater. The samples were stored for up to two months at room temperature using three different methods, and we observed changes in microbial communities over time. We analyzed clusters of changes in the microbial flora by observing fecal stock solutions and metagenome sequencing performed over time. In particular, we confirmed the profiling of alpha and beta diversity and microbial classification according to the differences in intestinal environment among individuals. We also confirmed that the microbial profile remained stable for two months and that the microbial profile did not change significantly over time. In addition, our results suggest the possibility of verifying microbial profiling even for long-term storage of a single sample. In conclusion, collecting fecal samples using a stock solution rather than freezing feces seems to be relatively reproducible and stable for GUT metagenome analysis. Therefore, stock solution tubes in intestinal microbial research can be used without problems.
Similar content being viewed by others
References
Bahl, M.I., Bergström, A., and Licht, T.R. 2012. Freezing fecal samples prior to DNA extraction affects the Firmicutes to Bacteroidetes ratio determined by downstream quantitative PCR analysis. FEMS Microbiol. Lett.329, 193–197.
Blum, H.E. 2017. The human microbiome: an emerging key player in health and disease. Arch. Clin. Biomed. Res.1, 85–95.
Bolyen, E., Rideout, J.R., Dillon, M.R., Bokulich, N.A., Abnet, C.C., Al-Ghalith, G.A., Alexander, H., Alm, E.J., Arumugam, M., Asnicar, F., et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol.37, 852–857.
Cho, I. and Blaser, M.J. 2012. The human microbiome: at the interface of health and disease. Nat. Rev. Genet.13, 260–270.
Collins, F.S., Morgan, M., and Patrinos, A. 2003. The human genome project: lessons from large-scale biology. Science300, 286–290.
Costea, P.I., Zeller, G., Sunagawa, S., Pelletier, E., Alberti, A., Levenez, F., Tramontano, M., Driessen, M., Hercog, R., Jung, F.E., et al. 2017. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol.35, 1069–1076.
David, L.A., Materna, A.C., Friedman, J., Campos-Baptista, M.I., Blackburn, M.C., Perrotta, A., Erdman, S.E., and Alm, E.J. 2014. Host lifestyle affects human microbiota on daily timescales. Genome Biol.15, R89.
Dominianni, C., Wu, J., Hayes, R.B., and Ahn, J. 2014. Comparison of methods for fecal microbiome biospecimen collection. BMC Microbiol.14, 103.
Fisher, R.A. 1936. The use of multiple measurements in taxonomic problems. Ann. Eugen.7, 179–188.
Fouhy, F., Deane, J., Rea, M.C., O’Sullivan, O., Ross, R.P., O’Callaghan, G., Plant, B.J., and Stanton, C. 2015. The effects of freezing on faecal microbiota as determined using MiSeq sequencing and culture-based investigations. PLoS One10, e0119355.
Hale, V.L., Tan, C.L., Knight, R., and Amato, K.R. 2015. Effect of preservation method on spider monkey (Ateles geoffroyi) fecal microbiota over 8 weeks. J. Microbiol. Methods113, 16–26.
Hale, V.L., Tan, C.L., Niu, K., Yang, Y., Cui, D., Zhao, H., Knight, R., and Amato, K.R. 2016. Effects of field conditions on fecal microbiota. J. Microbiol. Methods130, 180–188.
Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature486, 207–214.
Ihekweazu, F.D. and Versalovic, J. 2018. Development of the pediatric gut microbiome: impact on health and disease. Am. J. Med. Sci.356, 413–423.
Johnson, J.S., Spakowicz, D.J., Hong, B.Y., Petersen, L.M., Demkowicz, P., Chen, L., Leopold, S.R., Hanson, B.M., Agresta, H.O., Gerstein, M., et al. 2019. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun.10, 5029.
Knights, D., Silverberg, M.S., Weersma, R.K., Gevers, D., Dijkstra, G., Huang, H., Tyler, A.D., van Sommeren, S., Imhann, F., Stempak, J.M., et al. 2014. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med.6, 107.
Ley, R.E., Hamady, M., Lozupone, C., Turnbaugh, P.J., Ramey, R.R., Bircher, J.S., Schlegel, M.L., Tucker, T.A., Schrenzel, M.D., Knight, R., et al. 2008. Evolution of mammals and their gut microbes. Science320, 1647–1651.
Lynch, S.V. and Pedersen, O. 2016. The human intestinal microbiome in health and disease. N. Engl. J. Med.375, 2369–2379.
Nam, Y.D., Jung, M.J., Roh, S.W., Kim, M.S., and Bae, J.W. 2011. Comparative analysis of Korean human gut microbiota by barcoded pyrosequencing. PLoS One6, e22109.
Park, S.T. and Kim, J. 2016. Trends in next-generation sequencing and a new era for whole genome sequencing. Int. Neurourol. J.20, S76–S83.
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K.S., Manichanh, C., Nielsen, T., Pons, N., Levenez, F., Yamada, T., et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature464, 59–65.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., Peplies, J., and Glockner, F.O. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res.41, D590–D596.
Ravi, R.K., Walton, K., and Khosroheidari, M. 2018. MiSeq: A next generation sequencing platform for genomic analysis. Methods Mol. Biol.1706, 223–232.
Rodriguez, J.M., Murphy, K., Stanton, C., Ross, R.P., Kober, O.I., Juge, N., Avershina, E., Rudi, K., Narbad, A., Jenmalm, M.C., et al. 2015. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis.26, 26050.
Santiago, A., Panda, S., Mengels, G., Martinez, X., Azpiroz, F., Dore, J., Guarner, F., and Manichanh, C. 2014. Processing faecal samples: a step forward for standards in microbial community analysis. BMC Microbiol.14, 112.
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W.S., and Huttenhower, C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol.12, R60.
Sinha, R., Chen, J., Amir, A., Vogtmann, E., Shi, J., Inman, K.S., Flores, R., Sampson, J., Knight, R., and Chia, N. 2016. Collecting fecal samples for microbiome analyses in epidemiology studies. Cancer Epidemiol. Biomarkers Prev.25, 407–416.
Spor, A., Koren, O., and Ley, R. 2011. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol.9, 279–290.
Thursby, E. and Juge, N. 2017. Introduction to the human gut microbiota. Biochem. J.474, 1823–1836.
Tigchelaar, E.F., Bonder, M.J., Jankipersadsing, S.A., Fu, J., Wijmenga, C., and Zhernakova, A. 2016. Gut microbiota composition associated with stool consistency. Gut65, 540–542.
Turnbaugh, P.J., Hamady, M., Yatsunenko, T., Cantarel, B.L., Duncan, A., Ley, R.E., Sogin, M.L., Jones, W.J., Roe, B.A., Affourtit, J.P., et al. 2009. A core gut microbiome in obese and lean twins. Nature457, 480–484.
Vandeputte, D., Falony, G., Vieira-Silva, S., Tito, R.Y., Joossens, M., and Raes, J. 2016. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut65, 57–62.
Vogtmann, E., Chen, J., Amir, A., Shi, J., Abnet, C.C., Nelson, H., Knight, R., Chia, N., and Sinha, R. 2017. Comparison of collection methods for fecal samples in microbiome studies. Am. J. Epidemiol.185, 115–123.
Wang, Z., Zolnik, C.P., Qiu, Y., Usyk, M., Wang, T., Strickler, H.D., Isasi, C.R., Kaplan, R.C., Kurland, I.J., Qi, Q., et al. 2018. Comparison of fecal collection methods for microbiome and metabolomics studies. Front. Cell. Infect. Microbiol.8, 301.
Warnes, G.R., Bolker, B., Bonebakker, L., Genteman, R., Liaw, W., Lumley, T., Maechler, M., Magnusson A., Moeller, S., Schwartz, M., et al. 2015. gplots: Various R programming tools for plotting data. R package version 2.17.0.
Wu, G.D., Lewis, J.D., Hoffmann, C., Chen, Y.Y., Knight, R., Bittinger, K., Hwang, J., Chen, J., Berkowsky, R., Nessel, L., et al. 2010. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol.10, 206.
Young, V.B. 2017. The role of the microbiome in human health and disease: an introduction for clinicians. BMJ356, j831.
Acknowledgments
The authors thank CEO Samuel Hwang (Theragen Bio, Republic of Korea) for supporting the sample preparation, sequencing, and computing system for bioinformatics analyses.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that there are no conflicts of interest.
Additional information
Supplemental material for this article may be found at http://www.springerlink.com/content/120956.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Park, C., Yun, K.E., Chu, J.M. et al. Performance comparison of fecal preservative and stock solutions for gut microbiome storage at room temperature. J Microbiol. 58, 703–710 (2020). https://doi.org/10.1007/s12275-020-0092-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-020-0092-6