SmpB and tmRNA Orchestrate Purine Pathway for the Trimethoprim Resistance in Aeromonas veronii
- 1Key Laboratory of Tropical Biological Resources of Ministry of Education, School of Life and Pharmaceutical Science, Hainan University, Haikou, China
- 2Key Laboratory for Sustainable Utilization of Tropical Bioresource, College of Tropical Crops Hainan University, Haikou, China
Small protein B(SmpB) cooperates with transfer-messenger RNA (tmRNA) for trans-translation to ensure the quality control of protein synthesis in prokaryotes. Furthermore, they regulate cell metabolism separately. According to research, SmpB functions as a transcription factor, and tmRNA acts as a small RNA. Purine pathway has been reported to be related to trimethoprim resistance, including hypoxanthine synthesis, adenosine metabolism and guanosine metabolism. Another reason of drug tolerance is the efflux pump of the bacterium. In transcriptomic data, it was shown that the expression of some related enzymes in adenosine metabolism were raised significantly in smpB deletion strain than that of wild type, which led to the differential trimethoprim resistance of Aeromonas veronii (A. veronii). Furthermore, the metabolic products of adenosine AMP, cAMP, and deoxyadenosine were accumulated significantly. However, the expressions of the enzymes related to hypoxanthine synthesis and guanosine metabolism were elevated significantly in ssrA (small stable RNA, tmRNA) deletion strain, which eventually caused an augmented metabolic product xanthine. In addition, the deletion of ssrA also affected the significant downregulations of efflux pump acrA/acrB. The minimal inhibitory concentrations (MIC) were overall decreased after the trimethoprim treatment to the wild type, ΔsmpB and ΔssrA. And the difference in sensitivity between ΔsmpB and ΔssrA was evident. The MIC of ΔsmpB was descended significantly than those of wild type and ΔssrA in M9 medium supplemented with 1 mM adenosine, illustrating that the adenosine metabolism pathway was principally influenced by SmpB. Likewise, the strain ΔssrA conferred more sensitivity than wild type and ΔsmpB in M9 medium supplemented with 1mM guanosine. By overexpressing acrA/acrB, the tolerance to trimethoprim was partially recovered in ΔssrA. These results revealed that SmpB and tmRNA acted on different branches in purine metabolism, conferring the diverse trimethoprim resistance to A. veronii. This study suggests that the trans-translation system might be an effective target in clinical treatment of A. veronii and other multi-antibiotic resistance bacteria with trimethoprim.
Introduction
Aeromonas veronii (A. veronii) is a pathogen of aquatic animals and humans. It is widely found in freshwater and seawater with a strong pathogenic ability, causing fish skin ulceration, visceral hemorrhage, ascites, and other symptoms (Liu et al., 2016). Clinical studies have reported that A. veronii can also cause human gastroenteritis, endocarditis, bacteremia and other diseases (Aguilera-Arreola et al., 2007; Chuang et al., 2011). A. veronii is resistant to spectinomycin, gentamicin, streptomycin, and kanamycin (Zhang et al., 2019). Bacterial drug resistance makes it more difficult to control pathogens (Leal et al., 2019; El Mekes et al., 2020). Studies have shown that trans-translation system composed of SmpB (Small protein B) and tmRNA (transfer-messenger RNA) can release ribosomal block caused by translation errors, thus ensuring ribosome efficiency and protein synthesis (Keiler, 2015), which has an important impact on bacterial drug resistance (Venkataraman et al., 2014; Sharkey et al., 2016).
In addition to the trans-translation function of SmpB and tmRNA, they also play other roles in bacteria. As a transcription factor, SmpB participates in transcriptional regulation of bvgS and sRNA of A. veronii (Liu et al., 2015; Wang et al., 2019). SmpB also affects the transcription of sRNA and the tolerance to environmental stress (Wang et al., 2019). Analogously, tmRNA can act as sRNA and interact with crtMN mRNA to affect the synthesis of Staphylococcus aureus pigment (Liu et al., 2010). Results of the present study show that a double knockout of smpB and ssrA (tmRNA, small stable RNA) makes A. veronii more sensitive to trimethoprim.
It is reported that bacteria could develop acquired drug tolerance when treated with Trimethoprim (Ma et al., 2018). Trimethoprim affects the synthesis of nucleotides and purine metabolism of A. veronii by inhibiting dihydrofolate reductase (Sangurdekar et al., 2011). Purine metabolism also affects the bacterial tolerance to trimethoprim (Sah et al., 2015; Stepanek et al., 2016). This study explored the molecular mechanism of SmpB and tmRNA in the tolerance of A. veronii to trimethoprim, mainly through purine metabolism. In ΔsmpB, seven adenine related enzyme expression upregulated, and the content of adenosine increased. In ΔssrA, a large number of guanine related enzymes upregulated, the content guanine and hypoxanthine contents increased. SmpB and tmRNA affect two branches of purine metabolism, adenine synthesis and guanine synthesis, respectively. The sensitivity of A. veronii to trimethoprim increased with the addition of exogenous purine products. More different, ΔssrA efflux pump related gene expression was decreased, and the overexpression of acrAB made ΔssrA increased the resistance of trimethoprim. This study explained the reason for decreased MIC of trimethoprim, and the different influence between ΔsmpB and ΔssrA on purine pathway was also found.
Methods
Strains
The strain information was listed in Table 1. A. veronii was isolated from the diseased tissues of grass carp (Liu et al., 2015) and cultured with M9 Minimal Medium (M9) at 30°C, 150 r/min, and 50 ug/mL ampicillin. SmpB knockout strain and ssrA knockout strain was derived from the preserved strain in this laboratory (Liu et al., 2015). The smpB and ssrA double knockout strain (ΔsmpB_ssrA) and ssrA mutant complemented with acrAB sequence (ΔssrA::acrAB) were constructed in this study. E coli WM 3064 assisted the vector introduction into A. veronii for the construction of overexpressed strains and knockout strains. E coli WM3064 was Diaminopimelic acid (DAP) dependent and cultured in LB medium at 37°C with DAP concentration of 0.3mM. All of the medium composition belongs to inorganic salts. For the 5× stock: 64 g Na2HPO4, 15 g KH2PO4, 5 g NH4Cl, 2.5 g NaCl, and 1 liter of high-quality distilled water. Mix 1 ml of 1 M MgSO4.7H2O, 10 ml of 20% glucose, 100ul of 1M CaCl2 and 200mL of 5× stock solution and adjust to 1000ml with distilled H2O.
Plasmids
The construction vectors and primers used in this study are listed in Tables 1, 2. Suicide plasmid pRE112 was used to construct knockout vectors. And the assembly knockout vectors of smpB_ssrA sequence were constructed by double enzyme digestion and specific connection and were integrated with the genome under 6% sucrose to complete homologous recombination (Liu et al., 2015). Plasmid pBBR1MCS-2 was used to construct a vector that overexpressed efflux pump-related gene acrAB in A. veronii. DNA sequence acrA and acrB were introduced into the pBBR1MCs-2 plasmid, as well as the upstream and downstream primers needed for vector construction respectively. The plasmid was transferred into ΔssrA strain through affinity experiment to construct the overexpressed strain. The overexpressed strains were screened using plates containing both Kanamycin and Ampicillin resistance and verified by sequencing.
Minimum Inhibitory Concentration Test
Micro broth dilution method: Antibiotics were added to the sterile 96-well plates at final concentrations of 64, 32, 16, 8, 4, 2, 1, 0.5, 0.25, and 0.125 ug/mL. Then, 10-E6 CFU broth was added to each well to a final volume of 200 μL. The 96-well plate was sealed with parafilm and cultured at 30°C with shaking at 150 r/min for 24 h. The minimum drug concentration that completely inhibited the growth in the well was considered as the minimum inhibitory concentration. The experiment was repeated 3 times.
Transcriptomic Analysis
The cells were collected, lysed, and the sample RNA was extracted with phenol-chloroform. The concentration and quality of the RNA samples were tested with the Agilent 2100. DNase I was used to remove double-stranded DNA, and a Ribo-Zero Magnetic Kit was used to remove human RNA. Reverse transcription was performed with random primers and first strand cDNA which was used as a template to synthesize the second strand. The linker sequence was attached to the 3′ end of the cDNA fragment. The cDNA sequence was amplified with a primer cocktail, and the purified product was sequenced on a HiSeq Xten (Illumina, San Diego, CA, USA) platform. The sequencing depth was chain-specific sequencing for 2 Gb of clean data. HISAT was attempted for genome assembly, analysis of potential coding sequence analysis and to identify new transcripts that may be present. Differences in gene transcription levels between wild-type and knockout strains were analyzed by Bowtie 2, and RPKM was used to normalize gene expression levels. The results of gene expression were calculated using the Benjamini-Hochberg false discovery rate (FDR). The differentially expressed transcripts were tested for log-fold change and the p value was corrected with FDR <0.001. The results of the differentially expressed genes were analyzed using GO classification, and differential gene expression in the pathway was compared with the entire genomic background using hyper geometric analysis. Value of p < 0.05 is considered to be a differential metabolic pathway. The accession number of reference genome is NO. CP012504.1. GEO accession number is GSE120603, and the URL of accession website is https://submit.ncbi.nlm.nih.gov/subs/sra/SUB6133286. The DESeq.2 package in R was applied to estimate the fold changes and to perform other analysis.
Metabolomics Analysis
The non-target metabolomics and lipidomics detection platform (UHPLC-QTOF-MS) was applied to metabolomics for the detection of A. veronii samples. UHPLC-QTOF-MS includes Ultra-Performance Liquid Chromatography 1290UHPLC (Agilent), ACQUITY UPLC BEH Amide column 1.7 um, 2.1*100 mm (Waters) and High-Resolution Mass Spectrometry Triple TOF 6600 (AB Sciex). The original mass spectrum was converted to the mzXML format using Proteo Wizard software, and the peaks were identified using the R Programming Language package (Version 3.2) and self-built secondary mass spectrometry data. URL of accession website is: www.ebi.ac.uk/metabolights/MTBLS1411.
Statistical Analysis
Statistical data were analyzed using the Statistical Package for the Social Science (SPSS) version 20.0 (SPSS, Chicago, IL, USA) and GraphPad Prism version 8.0 (GraphPad, San Diego, CA, USA). The results are presented as the mean values of three independent experiments with standard deviation (SD) using one-way analysis of variance (ANOVA). Values of p < 0.05 or 0.01 were represented as significant or extremely significant, respectively.
Results
1. The trimethoprim tolerance of ΔsmpB_ssrA to was decreased than wild type A. veronii.
The minimum inhibitory concentration (MIC) was tested by trimethoprim gradient concentration. The trimethoprim MIC of ΔsmpB_ssrA was 2 ug/mL. Compared with wild type A. veronii, the strain of ΔsmpB_ssrA was more sensitive to trimethoprim (Figure 1A). In order to distinguish the function of SmpB and tmRNA on trimethoprim, ΔsmpB and ΔssrA strains were used in this study. The MIC data showed that ΔsmpB and ΔssrA were more sensitive than the wild type, which was still higher than ΔsmpB_ssrA. The MIC data of complementary strain was tested (Figure 1B). The changes between wild type and knockout strains were rescued by transferring their pBBR plasmids into the deficient strain. The growth curve was tested to prove that the MIC value was not influenced by the non-obvious difference on growth (Figure 1C) This phenomenon suggested that the functions of SmpB and tmRNA were different from trans-translation in the presence of trimethoprim.
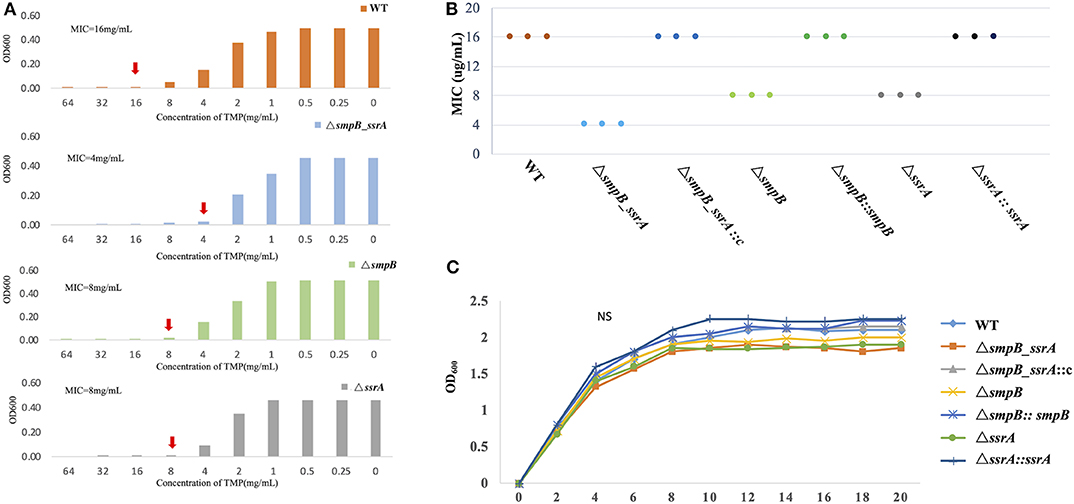
Figure 1. Minimum inhibitory concentration (MIC) value of trimethoprim. (A) The red arrow indicates the concentration of trimethoprim in M9 medium that prevented the growth of Aeromonas veronii. (B) The MIC of the complementary strain of the three mutants. (C) The growth curve of wild type A. veronii, ΔsmpB, ΔssrA, and ΔsmpB_ ssrA, and the complementary strains.
2. The difference between ΔsmpB and ΔssrA occurred at the transcriptional level.
In order to explore the differences on function of SmpB and tmRNA, the transcriptomic sequencing was taken to test the expression of stationary phage ΔsmpB and ΔssrA stains. The transcriptomic results showed that there was a big difference between ΔsmpB and ΔssrA (Figure 2A). As the expression of guanosine related enzymes in ΔssrA purine metabolism was significantly higher than that in wild type A. veronii. Simultaneously, the Expression of adenosine related enzymes in ΔsmpB purine metabolism was significantly higher than wild type A. veronii (Figure 2B).
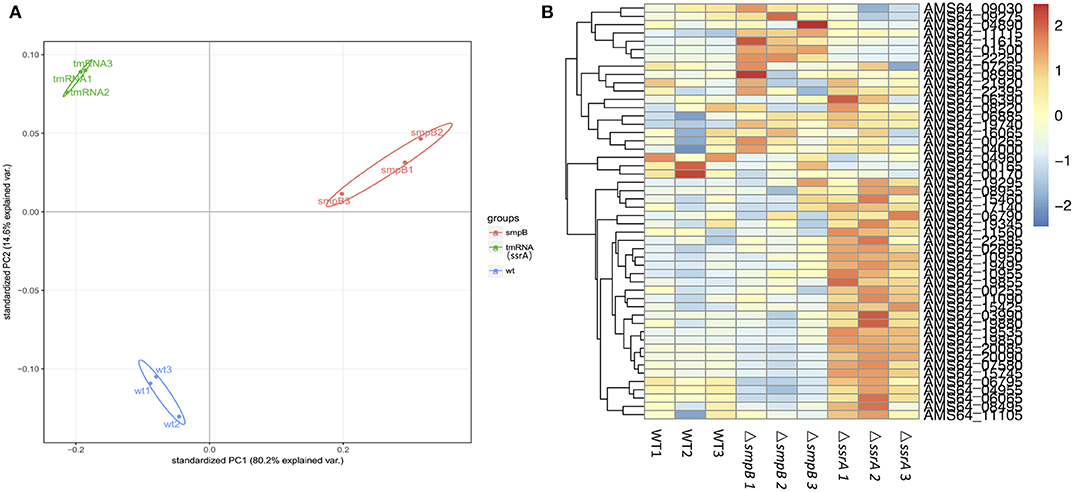
Figure 2. Transcriptomic data of Aeromonas veronii. (A) Principal component analysis (PCA) diagram of wild type, ΔsmpB and ΔssrA data. (B) Heat map of expression levels of genes involved in purine metabolism and efflux pump synthesis of wild type, ΔsmpB and ΔssrA strains. There were three samples of wild type, ΔsmpB and ΔssrA for transcriptome sequencing and analysing. The reads were mapped to reference genome with the accession NO. CP012504.1.
3. The content of purine metabolites in ΔsmpB and ΔssrA were different.
Metabonomic analysis of ΔsmpB and ΔssrA strains was done because of significant differences in the transcriptomic data. Metabolomics results showed that guanosine metabolites and adenosine metabolites showed up-regulation in the purine metabolic pathway separately in ΔsmpB and ΔssrA (Figure 3). The content of adenosine, AMP, cAMP, and deoxyadenosine in ΔsmpB were increased significantly. Whereas the contents of xanthine and guanosine in ΔssrA were significantly increased.
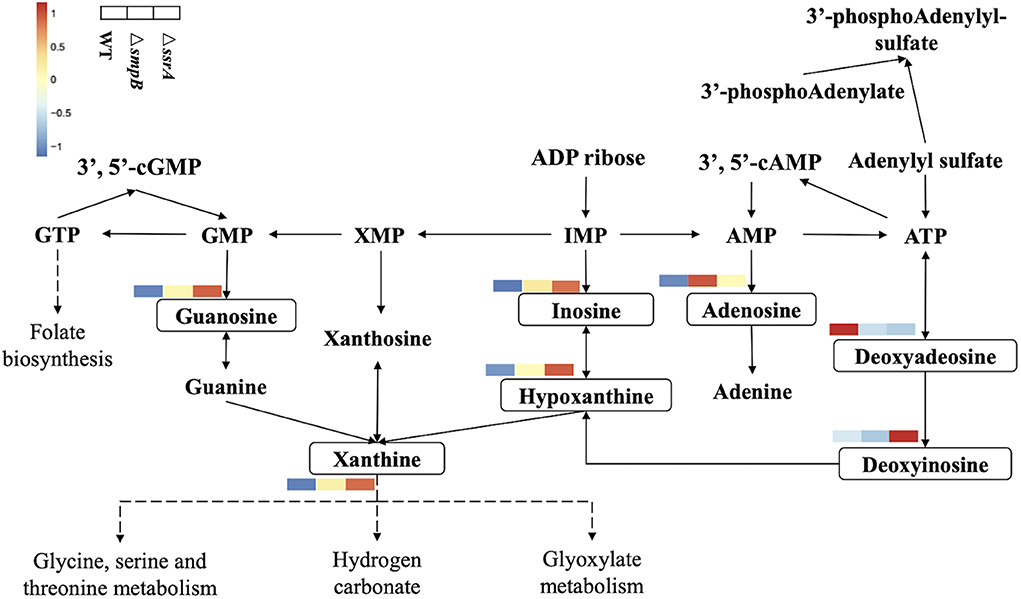
Figure 3. Simplified diagram of Aeromonas veronii purine metabolic pathway. The metabolites with a change between wild-type, ΔsmpB and ΔssrA strains were framed by solid line and presented by heat-map. Compounds with no significant difference in content of the three strains were not labeled.
4. Trimethoprim tolerance of ΔssrA was enhanced by acrAB overexpression.
The expression of efflux pump gene acrA and acrB in ΔssrA was downregulated, but no significant change in ΔsmpB (Figure 4A). The acrAB over-expression vector pBBR_acrAB, which was constructed with acrAB promoter, were introduced into ΔssrA. Trimethoprim MIC test results showed that the MIC of ΔssrA::acrAB similar to wild type strain (Figure 4B). The tolerance to trimethoprim is enhanced by over-expression of acrAB in ΔssrA. Under the condition of 1 mM adenosine in M9, the MIC of ΔssrA::acrAB strain increased to 8 mg/mL, which is lower than the wild type.
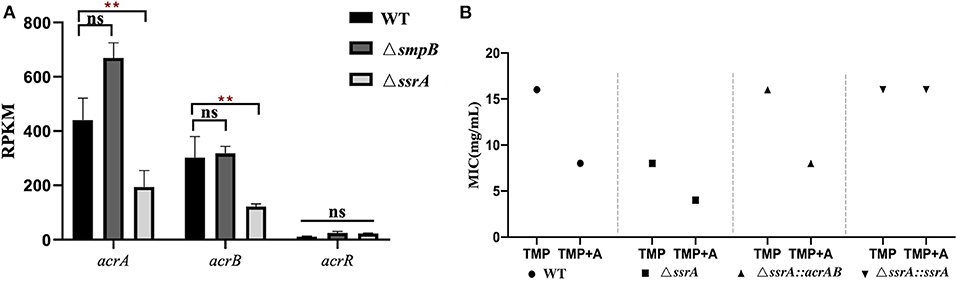
Figure 4. Trimethoprim tolerance of ΔssrA was enhanced by acrAB overexpression. (A) Expression of efflux pump related gene acrA and acrB in wild type, ΔsmpB and ΔssrA strains. (B) MIC value of wild type, ΔssrA strains affected by overexpression of acrAB. A represents the metabolite substrate 1 mM adenosine. **represents p < 0.01, and ns represents no significant difference.
5. The trimethoprim tolerance of ΔsmpB and ΔssrA was weakened by exogenous purine metabolites.
Excessive purine metabolites in M9 medium resulted in a decrease in MIC value, making the wildtype of A. veronii more sensitive to trimethoprim (Figure 5A, Supplementary Table 1). The MIC value of ΔsmpB was decreased by adding exogenous 1mM adenosine and 10mM ATP but was not changed by excessive guanosine (Figure 5B) and complementary strains (Supplementary Table 1). These results implied that the increase of adenosine in adenosine metabolic pathways could make ΔsmpB more sensitive to trimethoprim. Adding exogenous adenosine and guanosine, the MIC value of ΔssrA was decreased. Conversely, there was no change between the MIC of ΔssrA and the wild type cultivated by 10 mM ATP in M9 medium (Figure 5C). And 1mM guanosine in M9 medium had an effect on MIC value of ΔssrA. These results demonstrated that the differences in the regulation of purine metabolites led to differences in their tolerance to trimethoprim between ΔsmpB and ΔssrA.
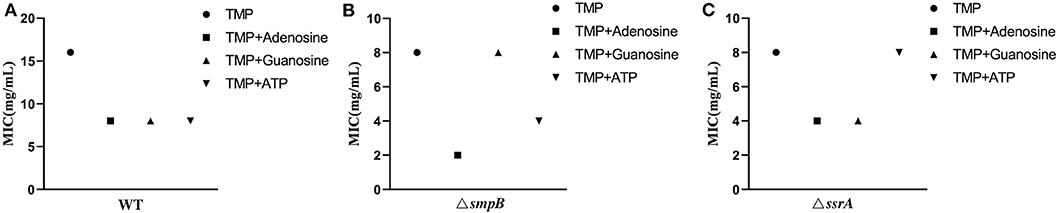
Figure 5. MIC value of trimethoprim under conditions of adenosine, guanosine and adenosine triphosphate. (A) MIC value of wild type with 1 mM adenosine, 1 mM guanosine, and 10 mM ATP in M9 medium. (B) MIC of ΔsmpB with 1 mM adenosine, 1 mM guanosine, and 10 mM ATP in M9 medium. (C) MIC of ΔssrA with 1 mM adenosine, 1 mM guanosine, and 10 mM ATP in M9 medium.
Discussion
Trimethoprim inhibits the synthesis of tetrahydrofolic acid, a carbon unit carrier, by affecting the activity of dihydrofolate reductase, thereby affecting the growth and metabolism of bacteria (Bertacine Dias et al., 2018). The products of purine metabolism will be reduced under the inadequate condition of one carbon unit (Sah et al., 2015). However, in the A. veronii metabonomic data, the decrease in folate metabolism and the increase in purine pathway-related enzymes were found to occur at the same time as the significant up-regulation of the purine metabolic pathway. The upregulation trend of ΔssrA was more sensitive to trimethoprim than that of ΔsmpB. The decrease of MIC value of trimethoprim to ΔsmpB and ΔssrA were not consistent with the double knockout strain. This indicated different functions of SmpB and tmRNA acting on the tolerance of trimethoprim. These results suggested that the reduced tolerance of trimethoprim in the double-knockout strain may be less relevant to the function of the trans-translation system.
ATP is known to be the most effective energy to physiological reactions. As an important products of purine metabolism, ATP was predicted to be beneficial to the growth of A. veronii. In this study, 10mM ATP led to a decrease in trimethoprim MIC value, suggesting that excessive ATP weakened the tolerance of A. veronii to trimethoprim. It was speculated that the heightened need of ATP increased the challenge to the one carbon unit synthesis of the organism. The role of efflux pump had been explored in drug resistance to fluoroquinolones in gram negative bacteria. AcrAB-TolC efflux system, which has a physiologic role of pumping out bile acids and fatty acids to lower their toxicity. The decreased expression of AcrAB also contributed to the accumulation of ATP. Therefore, it is speculated that ATP in a dynamic balance that It showed little influence upon the value of MIC.
Recent studies had been focused on the assistance of SmpB in binding to the stalled ribosome to release it (Keiler, 2015; Buskirk and Green, 2017). In ΔssrA, the expression of genes related to guanine synthesis was significantly upregulated and the mRNA of genes related to effusion pump was significantly decreased, which did not occur in ΔsmpB. On the contrary, the expression adenine-related enzymes of ΔsmpB were significantly increased, which were not repeated in ΔssrA. This indicates that SmpB and tmRNA have independent and non-interference functions.
The difference in tolerance between double knock out strain ΔsmpB_ssrA and single deletion strain ΔsmpB and ΔssrA indicated the distinctive functions. The transcriptional and metabolites data showed separate regulation of SmpB and tmRNA. The changes of ΔssrA were more significant. The trimethoprim MIC value results proved that the efflux pump AcrAB-TolC was affected by ssrA deletion. Therefore, this study demonstrated that SmpB and tmRNA act on different branches of purine metabolism, showing an effect on the tolerance to trimethoprim. A second collaboration between SmpB and tmRNA was completed by regulating purine metabolic pathway.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
ZL, YT, DH, and DW contributed the conception and design of the study. DW, HL, XM, and HT performed the statistical analysis. DW, WK, and ZL drafted the manuscript. All authors contributed to manuscript revision, read and approved the submitted version.
Funding
This work was supported by the Hainan Natural Science Foundation No. 319QN161 (to HL), the National Science and Technology Major Project No. 2019ZX08010004 (to ZL), and the grants from the National Natural Science Foundation of China Nos. 31560021 and 31772887 (to ZL) and 31860676 (to YT) and 31960027 (to XM), and the Priming Scientific Research Foundation of Hainan University No. KYQD (ZR) 1929 (to HL).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.00239/full#supplementary-material
References
Aguilera-Arreola, M. G., Hernandez-Rodriguez, C., Zuniga, G., Figueras, M. J., Garduno, R. A., and Castro-Escarpulli, G. (2007). Virulence potential and genetic diversity of Aeromonas caviae, Aeromonas veronii, and Aeromonas hydrophila clinical isolates from Mexico and Spain: a comparative study. Can. J. Microbiol. 53, 877–887. doi: 10.1139/W07-051
Bertacine Dias, M. V., Santos, J. C., Libreros-Zuniga, G. A., Ribeiro, J. A., and Chavez-Pacheco, S. M. (2018). Folate biosynthesis pathway: mechanisms and insights into drug design for infectious diseases. Future Med. Chem. 10, 935–959. doi: 10.4155/fmc-2017-0168
Buskirk, A. R., and Green, R. (2017). Ribosome pausing, arrest and rescue in bacteria and eukaryotes. Philos. Trans. R Soc. Lond. B Biol. Sci. 372:1716. doi: 10.1098/rstb.2016.0183
Chuang, H. C., Ho, Y. H., Lay, C. J., Wang, L. S., Tsai, Y. S., and Tsai, C. C. (2011). Different clinical characteristics among Aeromonas hydrophila, Aeromonas veronii biovar sobria and Aeromonas caviae monomicrobial bacteremia. J. Korean Med. Sci. 26, 1415–1420. doi: 10.3346/jkms.2011.26.11.1415
El Mekes, A., Zahlane, K., Ait Said, L., Tadlaoui Ouafi, A., and Barakate, M. (2020). The clinical and epidemiological risk factors of infections due to multi-drug resistant bacteria in an adult intensive care unit of University Hospital Center in Marrakesh-Morocco. J. Infect. Public Health. 13, 637–643. doi: 10.1016/j.jiph.2019.08.012
Keiler, K. C. (2015). Mechanisms of ribosome rescue in bacteria. Nat. Rev. Microbiol. 13, 285–297. doi: 10.1038/nrmicro3438
Leal, H. F., Azevedo, J., Silva, G. E. O., Amorim, A. M. L., de Roma, L. R. C., Arraes, A. C. P., et al. (2019). Bloodstream infections caused by multidrug-resistant gram-negative bacteria: epidemiological, clinical and microbiological features. BMC Infect Dis. 19:609. doi: 10.1186/s12879-019-4265-z
Liu, P., Chen, Y., Wang, D., Tang, Y., Tang, H., Song, H., et al. (2016). Genetic selection of peptide aptamers that interact and inhibit both small protein B and alternative ribosome-rescue factor A of Aeromonas veronii C4. Front. Microbiol. 7:1228. doi: 10.3389/fmicb.2016.01228
Liu, Y., Wu, N., Dong, J., Gao, Y., Zhang, X., Shao, N., et al. (2010). SsrA (tmRNA) acts as an antisense RNA to regulate Staphylococcus aureus pigment synthesis by base pairing with crtMN mRNA. FEBS Lett. 584, 4325–4329. doi: 10.1016/j.febslet.2010.09.024
Liu, Z., Liu, P., Liu, S., Song, H., Tang, H., and Hu, X. (2015). Small protein B upregulates sensor kinase bvgS expression in Aeromonas veronii. Front. Microbiol. 6:579. doi: 10.3389/fmicb.2015.00579
Ma, Q. P., Su, L., Liu, J. W., Yao, M. X., and Yuan, G. Y. (2018). Study on the association between drugresistance and gene mutations of the active efflux pump acrABtolC gene and its regulatory genes. Mol. Med. Rep. 17, 8228–8236. doi: 10.3892/mmr.2018.8916
Sah, S., Aluri, S., Rex, K., and Varshney, U. (2015). One-carbon metabolic pathway rewiring in Escherichia coli reveals an evolutionary advantage of 10-formyltetrahydrofolate synthetase (Fhs) in survival under hypoxia. J. Bacteriol. 197, 717–726. doi: 10.1128/JB.02365-14
Sangurdekar, D. P., Zhang, Z., and Khodursky, A. B. (2011). The association of DNA damage response and nucleotide level modulation with the antibacterial mechanism of the anti-folate drug trimethoprim. BMC Genomics 12:583. doi: 10.1186/1471-2164-12-583
Sharkey, L. K., Edwards, T. A., and O'Neill, A. J. (2016). ABC-F proteins mediate antibiotic resistance through ribosomal protection. MBio 7:e01975. doi: 10.1128/mBio.01975-15
Stepanek, J. J., Schäkermann, S., Wenzel, M., Prochnow, P., and Bandow, J. E. (2016). Purine biosynthesis is the bottleneck in trimethoprim-treatedBacillus subtilis. Proteomics Clin. Appl. 10, 1036–1048. doi: 10.1002/prca.201600039
Venkataraman, K., Guja, K. E., Garcia-Diaz, M., and Karzai, A. W. (2014). Non-stop mRNA decay: a special attribute of trans-translation mediated ribosome rescue. Front. Microbiol. 5:93. doi: 10.3389/fmicb.2014.00093
Wang, D., Li, H., Ma, X., Tang, Y., Tang, H., Hu, X., et al. (2019). Small RNA AvrA regulates IscR to increase the stress tolerances in SmpB deficiency of Aeromonas veronii. Front. Cell Infect. Microbiol. 9:142. doi: 10.3389/fcimb.2019.00142
Keywords: Aeromonas veronii, trimethoprim, SmpB, tmRNA, purine metabolism
Citation: Wang D, Li H, Khan WU, Ma X, Tang H, Tang Y, Huang D and Liu Z (2020) SmpB and tmRNA Orchestrate Purine Pathway for the Trimethoprim Resistance in Aeromonas veronii. Front. Cell. Infect. Microbiol. 10:239. doi: 10.3389/fcimb.2020.00239
Received: 27 December 2019; Accepted: 27 April 2020;
Published: 25 May 2020.
Edited by:
Bo Peng, Sun Yat-sen University, ChinaReviewed by:
Vijayakumar Sekar, Shandong University, Weihai, ChinaHyouta Himeno, Hirosaki University, Japan
Copyright © 2020 Wang, Li, Khan, Ma, Tang, Tang, Huang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanqiong Tang, tyq@126.com; Zhu Liu, liuzhuabel@163.com
 Dan Wang
Dan Wang Hong Li
Hong Li Wasi Ullah Khan2
Wasi Ullah Khan2  Zhu Liu
Zhu Liu