Drosophila as a Model Organism in Host–Pathogen Interaction Studies
- Biomedical Sciences Department, College of Health Sciences, QU Health, Qatar University, Doha, Qatar
Owing to the genetic similarities and conserved pathways between a fruit fly and mammals, the use of the Drosophila model as a platform to unveil novel mechanisms of infection and disease progression has been justified and widely instigated. Gaining proper insight into host–pathogen interactions and identifying chief factors involved in host defense and pathogen virulence in Drosophila serves as a foundation to establish novel strategies for infectious disease prevention and control in higher organisms, including humans.
Introduction
Drosophila, a chief tool in contemporary genetic studies, became one of the most powerful model organisms widely used in scientific explorations. The versatility, low cost, short life cycle, well-characterized genome, and feasibility of genetic manipulation made the fruit fly an indispensable model organism for basic research. The modern era of Drosophila research initially took off when the fly was deployed in developmental biology, particularly in fly embryo studies to identify novel genes involved in development (Nusslein-Volhard and Wieschaus, 1980). Further studies conducted in Drosophila have contributed to novel groundbreaking findings that allowed the identification of fundamental components of different pathways conserved between the fruit fly and higher mammalian organisms, including humans. Recently, Drosophila gained great popularity in host–pathogen interaction and infectious disease control studies due to several reasons, many of which were attributed to evolutionary conserved features in both Drosophila and vertebrates including innate immune cascades, signal transduction pathways, and transcriptional regulators. The fruit fly surprisingly serves as a host for a diversity of pathogens and could be readily infected with these pathogens naturally or in an experimental setting. The existence of a wide array of molecular and genetic tools that allow gene manipulation in specific cells/tissues in the fly also favors its use in host–pathogen interaction studies. Genetic and genome-wide RNAi screens in either intact flies or cell lines have identified a wide array of host effector molecules and pathways involved in host defense against invading pathogens. Reciprocally, flies can be used to screen for pathogen-virulence factors. The fly's GAL4-UAS transactivation system (Brand and Perrimon, 1993) allows the direct expression of transgenes encoding host or pathogen proteins in a cell-type-specific manner in vivo. Also, the fly's LexA transcriptional system, allows combinatorial gene expression in a distinct or overlapping fashion in vivo (Pfeiffer et al., 2010; Yagi et al., 2010), opening up for the feasibility of conducting epistasis analysis and revealing a role of specific genes in regulating cellular processes and pathways. Such experiments are difficult to be conducted in higher model organisms including mammals, advocating the use of Drosophila in host–pathogen interaction studies. Like all invertebrates, Drosophila lacks an adaptive immune response and relies exclusively on innate immunity with both its humoral and cellular arms to fight off invading pathogens. These innate immune responses mainly include production of antimicrobial peptides (AMPs) and anti-pathogenic factors through core signaling pathways (Toll, IMD, and JAK/STAT), anti-viral response through the RNA interference (RNAi) pathway, and pathogen immobilization through phagocytosis, encapsulation, and melanization (Agaisse and Perrimon, 2004; Akira et al., 2006; Govind, 2008). In this review, we provide an overview of the use of Drosophila in host–pathogen interaction studies and highlight the role of the fly's innate immune system in pathogen control. We also recapitulate a broad spectrum of host defense and pathogen virulence factors identified in Drosophila-pathogen studies and involved in microbial control and disease progression.
Host Defense Factors
Drosophila is considered a significant model organism in studying host–pathogen interactions (Figure 1). The establishment of the D. melanogaster whole genome sequence in 2000 (Adams et al., 2000) paved the way for adapting existing high-throughput RNAi screening methodologies in Drosophila cell lines to study gene function and identify specific gene targets and immune-associated components and modulators (Ueda, 2001; Kiger et al., 2003). Combining the findings of high-throughput RNAi screens with classical genetic methods and in vivo fly studies enabled the identification of humoral and cell-mediated host defense factors against a wide array of intracellular and extracellular pathogens (Cherry, 2008; Bier and Guichard, 2012).
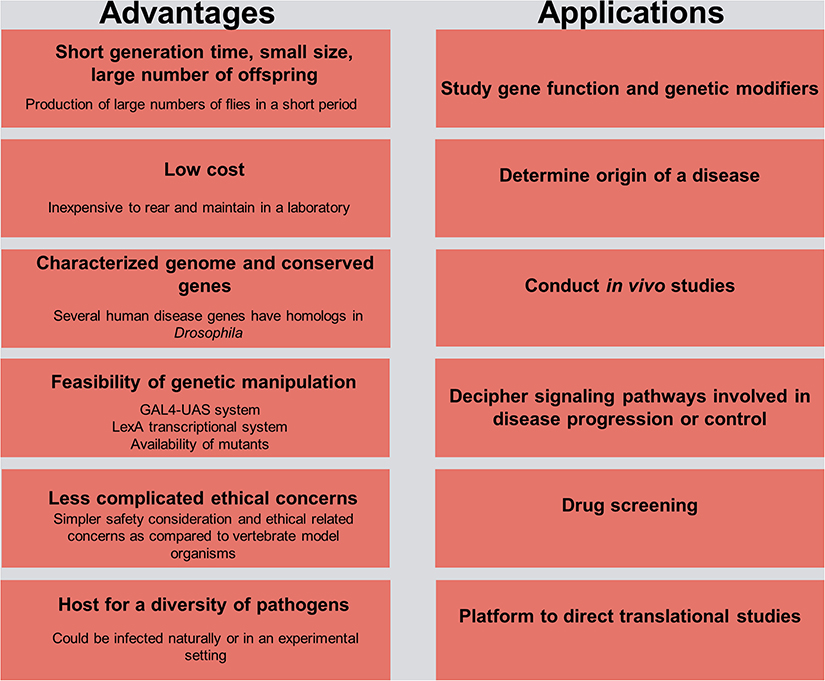
Figure 1. Advantages and practical applications in Drosophila for host–pathogen interaction studies. The left side of the figure delineates the advantages of using the fruit fly model organism in research, and the right side outlines its use as a platform for understanding the etiology of a disease and the potential means of controlling it.
Humoral Host Defense
Humoral innate immune responses in Drosophila mainly include production of AMPs and anti-pathogenic factors through Toll, IMD, and JAK/STAT signaling pathways. The primarily role attributed to the Toll pathway was its involvement in Drosophila embryonic development (Nusslein-Volhard and Wieschaus, 1980). In 1995, Hultmark et al. introduced Toll (Toll-1) as a potent immune activator in fruit fly cell lines (Rosetto et al., 1995). Since then, the Toll pathway was shown to be implicated in immune defense against an array of pathogens. Unlike the mammalian Toll pathway, the activation of Drosophila Toll signaling is not initiated by direct interaction with microbial determinants, but rather by the cleaved form of spätzle, a cytokine-like molecule that is thought to be processed by secreted serine proteases (SPs) and spätzle-processing enzyme (SPE). SPs and SPE are regulated by several pathogen recognition receptors (PRRs) including peptidoglycan recognition protein SA (PGRP-SA), PGRP-SD, Gram-negative binding protein 1 (GNBP1), and GNBP3 (Gottar et al., 2006). To avoid exaggerated immunity, the activation of the Toll pathway is generally tightly regulated. Upregulation of Spn1, a member of the serpin superfamily protease inhibitors located upstream of SPE, for example, contributes to the Toll pathway inactivation and to a downregulation in the expression of AMPs, mainly Drosomycin. Fungal-infected Spn1 null mutants exhibit an up-regulation in Drosomycin (Fullaondo et al., 2011). ModSP, a modular serine protease, activates the Toll pathway to culminate in AMP production. ModSP mutant flies challenged with either gram-positive bacteria (Enterococcus faecalis or Listeria monocytogenes) or fungal species (Candida albicans) succumb to death-associated reduction in AMP gene expression (Buchon et al., 2009a). In addition to its well-defined role against fungal and gram-positive bacteria, Oh et al. reported a role of the Toll pathway in defense against acid-fast mycobacteria. Mycobacterium abscessus, a non-tuberculous mycobacteria in humans, colonizes the gut of D. melanogaster and induces predominant expression of Drosomycin upon Toll pathway activation (Oh et al., 2013). Strikingly, Gottar et al. identified a pathway that acts jointly with GNBP3 to activate the Toll pathway upon fungal infection. PR1, a C. albicans virulence factor, activates Toll signaling by promoting the proteolytic cleavage and maturation of the Persephone protease (PSH). This finding indicates that the detection of fungal infection in Drosophila is dependent on both the recognition of foreign fungal invariant patterns and on tracking the consequence of virulence elements on the infected host (Gottar et al., 2006). Interestingly, and although both GNBP3 and PSH-dependent pathway are also required for Toll pathway activation upon Candida glabrata infection, only GNBP3 mutants are susceptible to Candida glabrata infection, implicating that the downstream effector mechanisms like AMP production and melanization activated against different fungal infections may not be the same (Chamilos et al., 2010; Quintin et al., 2013). Several studies have also employed D. melanogaster as a model organism to characterize anti-viral Toll immunity. The Toll pathway was shown to play a role in efficiently inhibiting Drosophila X viral (DXV) replication. Interestingly, the levels of Drosophila AMP genes induced in response to DXV infection were similar to those reported during Escherichia coli infection (Zambon et al., 2005). Extracellular virions, which were first discovered in Drosophila, and currently in metazoans, are also recognized by Toll-like receptors located on cell surfaces and inside endo-lysosomal compartments (Medzhitov, 2001).
Recently, the impact of post-translational modifications on modulating Toll signaling has been also studied in fruit flies. Such modifications were shown to change the localization and trafficking of the protein in a cell, enhance or inhibit the protein activity, and/or alter the protein's ability to bind to protein signaling partners. The Drosophila Ubc9/Lwr enzyme, for example, affects Toll signaling by stimulating the sumoylation of the Dorsal transcription factor (Schmidt, 2014). Likewise, β-arrestin Kurtz (Krz) regulates Toll signaling via protein sumoylation by interacting with the SUMO protease Ulp1. Krz or Ulp1 Drosophila larval mutants exhibit inflammation-like phenotypes characterized by elevation in lamellocyte production, formation of melanotic tumors, accumulation of transcriptional effectors (Dorsal and Dif) of the Toll pathway, and increased expression of anti-microbial peptides (Drosomycin). Interestingly, loss of function of these two genes reveal a dose-dependent sensitive and synergistic response, suggesting that they belong to the same signaling pathway (Anjum et al., 2013). Moreover, Pellinos, a family of E3 ubiquitin ligases, were shown to also regulate Toll signaling by catalyzing the K63-linked polyubiquitination of Pelle, an IL-1 receptor-associated kinase homolog in Drosophila (Medvedev et al., 2015). Genome-wide screening studies of the Toll pathway also identified novel immune-associated components and regulators including the Deformed Epidermal Auto-regulatory Factor 1 (DEAF1) transcription factor as an essential component for the expression of the Toll target AMP Drosomycin (Kuttenkeuler et al., 2010).
Similar to the Toll pathway, the Drosophila IMD pathway, which is mainly directed against gram-negative pathogens, plays a fundamental role in humoral immunity through AMP production and pathogen clearance. IMD mutant flies, for example, are sensitive to Vibrio cholerae infection (Wang et al., 2013; Kamareddine et al., 2018a), while those with a gain-of-function mutation exhibit resistance, plausibly by lowering the virulence effect of the cholera toxin via increasing the rate of intestinal stem cell division (Wang et al., 2013). DreddD55 IMD mutant flies infected with Xenorhabdus nematophila and Photorhabdus luminescens nemato-bacterial composites also fail to survive infection compared to Dif1 Toll mutants and wild-type infected flies, albeit the 24 h priming with non-pathogenic E. coli prior to X. nematophila and P. luminescens infection. These findings advocate the notion that X. nematophila and P. luminescens pathogens target components of the IMD pathway, despite AMPs synthesis triggered by the nemato-bacterial composite infection (Aymeric et al., 2010). Apart from its well-defined role against gram-negative bacteria, recent studies have also highlighted a role of IMD signaling in defense against fungal and gram-positive bacterial infection (De Gregorio et al., 2002a; Hedengren-Olcott et al., 2004; Pham et al., 2007; Dionne and Schneider, 2008; Costa et al., 2009). Interestingly, non-canonical AMP-independent IMD immunity have been also shown to be crucial in the Drosophila gut defense system. Hori et al. reported that IMD mutant flies are sucseptible to Staphylococcus aureus oral infections and revealed a role of the IMD pathway in clearance of S. aureus from the fly gut (Hori et al., 2018). In alliance with this distinctive role in gut immunity, the IMD pathway was also shown to control gut homeostatic balance in a microbiota-dependent–infection-independent context. The gut flora, which induces IMD signaling activation, significantly affects the midgut transcriptome and promotes the expression of key genes involved in host physiology. A study by Kamareddine et al. revealed that IMD signaling in enteroendocrine cells activated by the intestinal microbiota acetate metabolite regulates the expression of the tachykinin peptide hormone, promoting metabolic homeostasis in the host. Both germ-free flies and IMD mutant flies were shown to behave similarly by exhibiting developmental retardation, disrupted lipid metabolism, and a status of inactive insulin signaling (Kamareddine et al., 2018a). Owing to the fact that humoral immunity in Drosophila is chiefly mediated by AMP production by fat body cells, particular attention has been also given to our understanding of IMD signaling in the fat body. A study by Tsichritzis et al. (2007) revealed that the deubiquitinase Cylindromatosis (CYLD) inhibits NF-κB signaling and downregulates the IMD response. Although CYLD mutant flies exhibit an increase in AMP expression, particularly those with prior infections, yet these mutants succumb to death significantly faster than controls upon E. coli infection. Although the target of CYLD in the IMD pathway remains uncharacterized, this poor survival rate of CYLD-deficient flies could be attributed to an alteration in the structure and function of fat body cells, as CYLD regulates homeostatic balance in these cells. Interestingly, several factors that affect physiology and development in a host also affect Toll and IMD signaling through manipulating fat body maturation. The induction of Diptericin expression in larvae, for example, is affected by age and is dependent on the presence of the ecdysone molting hormone. A mutation affecting the metabolism of ecdysone could indirectly affect the immune status of a host (Meister and Richards, 1996; Ligoxygakis et al., 2002a). It is worth noting here that signaling mechanisms between the gut and the fat body contribute to the regulation of systemic immune responses in the host (Lemaitre and Hoffmann, 2007). Upon oral infection, Ecc15 and P. entomophila can colonize and multiply in the fly gut, triggering strong systemic immunity, without a need for those bacterial species to cross the wall of the gut (Vodovar et al., 2005; Acosta Muniz et al., 2007).
Since the IMD pathway is similar to the mammalian tumor necrosis factor receptor (TNFR) pathway (Leulier et al., 2000; Costa et al., 2009), which plays a critical role in infectious disease control particularly against viral infections (Herbein and O'Brien, 2000), several studies deployed Drosophila as a model organism to gain further insight into the role of IMD signaling in anti-viral immunity. The cricket paralysis virus (CrPV), an RNA virus that infects a wide range of insect hosts, displays increased virulence with higher viral loads in IMD mutant flies (Costa et al., 2009). Interestingly, IMD signaling-mediated anti-CrPV immunity seems to be also AMP independent. Similar to CrPV infection, sindbis viral replication increases in IMD mutant flies (Avadhanula et al., 2009). Moreover, knocking down the peptidoglycan recognition protein-LC (PGRP-LC), a membrane associated IMD pathway receptor, in Drosophila S2 cells also promotes an increase in the genome copy number of the sigma virus and causes a significant up-regulation in the expression of the L gene compared to other viral genes (Liao et al., 2019). An anti-microbial RNAi signaling screen performed by Foley et al. publicized different classes of negative and positive gene regulators of IMD signaling including those that enhance response to peptidoglycan stimulation (46 EDRi genes), and others that constitutively activate NF-kB in the absence of LPS induction (26 CDRi genes) (Blandin et al., 2004). Further screens identified additional IMD positive regulators including Iap2 and TAB (Garver et al., 2006; Kawai and Akira, 2006). Similar to the studies that have been conducted on the Toll pathway, the impact of post-translational modifications on regulating IMD signaling has been also recently deliberated in fruit flies. SP36/Scny was shown to negatively regulate IMD signaling transduction by hydrolyzing UbK63, a key player in IMD ubiquitination (Thevenon et al., 2009). Similarly, USPs were also shown to regulate IMD immune signaling. USP2, for instance, deubiquitinates Imd, promoting its degradation (Engel et al., 2014).
The JAK/STAT signaling pathway, which controls various biological processes and tissue hemostasis in both mammals and invertebrates, also contributes to humoral immunity in a host. It is mainly activated upon microbial infection and/or cellular damage induced by stress response/pathogen infection, and culminates in the production of regulatory molecules, anti-viral agents, and anti-bacterial agents including AMPs. Cell damage induced by Serratia marcescens and Erwinia carotovora infection in Drosophila for example induces JAK/STAT signaling (Buchon et al., 2009b; Cronin et al., 2009) and activates a gut-specific defense machinery characterized by the expression of a subset of AMPs including the Drosomycin-like peptide (dro3). This activation, which is pathogen specific and triggered by cell damage caused by bacterial infection rather than by the bacteria itself (Buchon et al., 2009c), is particularly important in maintaining gut homeostasis by controlling epithelial cell proliferation and renewal in response to bacterial infection (Buchon et al., 2009b; Jiang et al., 2009). In the absence of infection, the indigenous gut flora triggers the expression of hopTum−l or upd3, which is generally adequate to induce intestinal stem cell progeny differentiation and gut regeneration through JAK/STAT and JNK signaling (Buchon et al., 2009b). Moreover, global gene expression analysis of Drosophila gut tissues to oral Erwinia carotovora infection revealed an important contribution of IMD and JAK/STAT pathways, but not the Toll pathway, to the regulation of gut immune responses (Buchon et al., 2009c). Although the intricate contribution of JAK/STAT signaling to cellular immunity has not been fully understood, it has been thought to be involved in cellular responses like hemocyte proliferation and differentiation (Agaisse and Perrimon, 2004). Recently, Yang et al., reported a role of JAK/STAT signaling in parasitoid egg wasp encapsulation in infected Drosophila larvae (Yang et al., 2015). Several studies addressing the role of JAK/STAT pathway in anti-viral immunity also revealed that the expression of “traditional” JAK/STAT pathway target genes including upd2, upd3, and TotM, is induced by many viral species including vesicular stomatitis virus, Flock House virus, and Drosophila X virus (Kemp et al., 2013; Myllymaki and Ramet, 2014). Likewise, the Drosophila C virus infection triggers the expression of several genes like virus-induced RNA-1 (vir-1). Many of these induced genes enclose STAT binding sites in their promoter regions, and their activation is therefore dependent on JAK/STAT signaling. The JAK tyrosine kinase Hopscotch (Hop) was also shown to be involved in controlling Drosophila C virus loads and to participate in inducing the expression of some virus-regulated genes. Deficiencies in JAK/STAT signaling increases Drosophila C virus load and exhibits high mortality rates in infected flies (Dostert et al., 2005). Although double-stranded RNA (dsRNA) itself does not induce viral response in Drosophila, recent studies have shown that recognizing virus-derived dsRNA through the amino terminal DExD/H-box helicase domain of Dicer-2 promotes the expression of the vago-secreted protein (Paradkar et al., 2012) that plays an antiviral role against Drosophila C virus infection (Paradkar et al., 2012). Interestingly, vago seems to induce the JAK/STAT pathway through a Dome-independent mechanism, signifying the existence of an alternative receptor that is yet to be determined. This finding provides a conceivable role of vago in connecting both RNAi and JAK/STAT signaling pathways, suggesting that vago, which is thought to be insect specific, could serve as a cytokine and functionally relate to the mammalian interferon system (Paradkar et al., 2012). By comparing RNA interference (detailed in the section below) with JAK/STAT anti-viral immunity, however, RNAi interference epitomizes an effectual antiviral machinery that operates against an array of RNA and DNA viruses, unlike the antiviral contribution of JAK/STAT signaling, which seems to be more species specific (Kemp et al., 2013) (Figure 2).
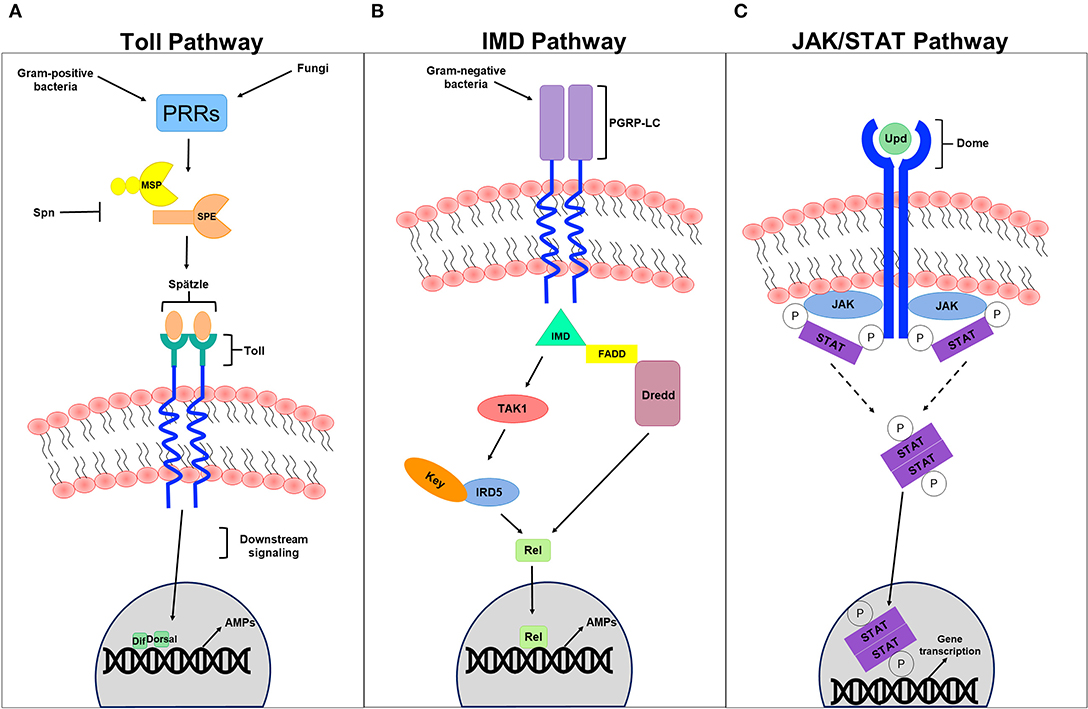
Figure 2. Humoral innate immune signaling pathways. (A) Represents a schematic diagram of the Toll pathway. Gram-positive bacteria and fungi recognized by pathogen recognition receptors (PRRs) trigger the activation of this pathway. Modular serine protease (MSP) and spätzle-processing enzyme (SPE), which are regulated by several PRRs are thought to process the cleavage of the spätzle ligand into a mature spätzle that binds to the Toll receptor, initiating downstream signaling pathway that culminates in the translocation of the NFB-like transcription factors Dif and/or Dorsal into the nucleus, promoting the expression to antimicrobial peptides (AMPs) in response to infection. Serpin (Spn) tightly regulates the primary steps of this pathway to avoid exaggerated immunity. (B) Represents a schematic diagram of the IMD pathway. Gram-negative bacteria recognized by receptors of the IMD pathways like the peptidoglycan recognition protein-LC (PGRP-LC) trigger the pathway activation, promoting the formation of the IMD, FADD, and Dredd (caspase 8 homolog) complex. This in turn activates Dredd, which is thought to be involved in the cleavage of the NFB-like transcription factors Relish (Rel). This formed complex also activates Tak1 (MAP3 kinase) and the IKK complex (IRD5 and key) to phosphorylate Rel. Once translocated into the nucleus, Rel promotes the expression to AMPs in response to invading pathogens. (C) Represents a schematic diagram of the JAK/STAT pathway. The UPD ligand binds to the DOME receptor leading to its activation. The phosphorylation of JAK and DOME create docking puts for STATs recruited to the formed complex. STATs themselves become phosphorylated generating an active dimer that translocates to the nucleus, promoting effector gene expression.
Cell-Mediated Host Defense
Phagocytosis, which is involved in ingesting apoptotic debris and destroying foreign pathogens by hemocytes (plasmatocytes, crystal cells, and lamellocytes), represents a fundamental mean of maintaining tissue homeostasis (Lemaitre and Hoffmann, 2007). Various receptors and key players chiefly involved in the phagocytic process have been identified in Drosophila. Pearson et al. revealed that the Drosophila scavenger receptor C1 (SR-CI), which has a broad polyanionic ligand-binding specificity similar to the mammalian class A macrophage-specific scavenger receptor (SR-A), exhibits great affinity and saturable binding of 125I-labeled acetylated low-density lipoprotein when expressed in mammalian cells (Pearson et al., 1995). Cuttell, et al. highlighted a previously uncharacterized role of the CED1/6/7 pathway in phagocytosis, by demonstrating that Draper (a CED-1homolog that belongs to the CED1/6/7 pathway)-mediated phagocytosis requires the Drosophila Junctophilin protein, Undertaker (UTA), and is linked to Ca2+ homeostasis (Cuttell et al., 2008). Additionally, Kocks et al. identified a role of Eater, an EGF-like repeat transmembrane receptor of the Nimrod family present on Drosophila hemocytes, in bacterial phagocytosis (Kocks et al., 2005). Likewise, Bretscher et al., uncovered the contribution of Eater in hemocyte localization, attachment, and adhesion, and in efficient phagocytosis of gram-positive (Staphylococcus aureus, Staphylococcus epidermidis, Micrococcus luteus), but not gram-negative (Escherichia coli and Serratia marcescens) bacteria (Bretscher et al., 2015). The intergin βν phagocytic receptor was also shown to be involved in defense against septic but not oral S. aureus infection in Drososphila (Shiratsuchi et al., 2012). Studies in Drosophila S2 cells in turn identified a role of PGRP-LC in phagocytosis of gram-negative (E. coli), but not gram-positive bacteria (Ramet et al., 2002). Apart from its scavenger function, PGRP-SC1 was also shown to act as an opsonin, and therefore, contribute to bacterial phagocytosis (Garver et al., 2006). Several thioester proteins (TEPs) identified in different insect species including Anopheles gambiae, also act as a bona fide opsonin to promote gram-positive and gram-negative bacterial phagocytosis (Levashina et al., 2001). In Drosophila, functional data publicized a role of several fruit fly TEPs, including TEP2, TEP3, and TEP6 in binding to several pathogens including E. coli, S. aureus, and C. albicans, respectively (Stroschein-Stevenson et al., 2006). Interestingly, Croquemort (CRQ), a CD36-related receptor that is exclusively expressed on macrophages in Drosophila embryo, was shown to be required for effectual phagocytosis of apoptotic corpses, but is not necessary for bacterial engulfment (Franc et al., 1999). Several screens identified cellular mediators of phagocytosis. Among those genes are four transcription factors, one of which encodes the GATA-factor Serpent, a chief regulator of hematopoesis in flies. Complimentary expression profile studies identified 45 genes, including the SR-C1 scavenger receptor gene that is down-regulated upon Serpent depletion (Meister and Tuschl, 2004; Haasnoot and Berkhout, 2006). RNAi against these Serpent-dependent genes further identified Eater and Nimrod phagocytic receptors (Miyano-Kurosaki and Takaku, 2006). Given that various classes of entry receptors plausibly facilitate the uptake of different microbes, although overlying and repetitive specificities do exist occasionally, many screen studies are usually done following specific microbial infections. Stroschein-Stevenson et al. identified 184 genes essential for efficient fungal uptake using Candida-infected phagocytic S2 cells. Among those genes is the macroglobulin-related protein (Mcr), which specifically opsonize Candida, unlike TEP2 and TEP3 that are needed for opsonization and efficient uptake of E. coli and S. aureus, respectively (Stroschein-Stevenson et al., 2006). Another screen following Mycobacteria fortuitum infection revealed 54 genes including the novel class B scavenger receptor peste (Li et al., 2002) that is essential for Mycobacteria fortuitum and Listeria monocytogenes, but not for E. coli and S. aureus uptake in S2 cells (Galiana-Arnoux et al., 2006). Genome-wide RNA interference screening was put forth to introduce host factors that block intracellular bacterial pathogenesis using cells. Interestingly, comparative studies of host defense genes involved in hindering bacterial pathogenesis revealed that some host factors have general inhibitory roles in intracellular pathogenesis, while others specifically affect the mechanistic ability of certain bacterial species to access the host (Agaisse et al., 2005). Rab7, CG8743, and the ESCRT machinery, for instance, represent unique vulnerability factors of the host cell, as manipulating any of these factors alone no longer constrains the growth of the non-pathogen Mycobacterium smegmatis in Drosophila (Yang et al., 2015). A similar study on Mycobacterium marinum also identified the lysosomal enzyme beta-hexosaminidase as an imperative factor in modulating mycobacterial growth. Remarkably, this bactericidal activity of β-hexosaminidase seems to be Mycobacterium marinum specific, as it is not involved in constraining the growth of other bacterial species like Salmonella typhimurium and Listeria monocytogenes (Koo et al., 2008).
Encapsulation is a another cellular response that is devoted to eliminate pathogens by forming hemocytic capsules around foreign bodies that are outsized to be phagocytozed (Kounatidis and Ligoxygakis, 2012). In Drosophila, cellular encapsulation happens in three stages. During the first stage, hemocytes, plausibly through their surface receptors, primarily recognize the parasitoid egg as a non-self. This recognition further promotes changes in the hemocyte cell surface membrane, exposing hidden molecules and presumably triggering downstream signaling (Nappi et al., 1991, 2000). During the second stage of encapsulation, the number of circulating hemocytes increases for a short term, and lamellocytes differentiate from plasmatocytes (Rizki and Rizki, 1990). Plasmatocytes account for more than 90% of all mature larval hemocytes and are involved in the phagocytic elimination of pathogenic microorganisms and dead cells (Lemaitre and Hoffmann, 2007). Activated lamellocytes, which are only present in larvae and whose expression is mainly induced upon infection, traffic to the parasitoid egg, flatten, and attach to the egg and to each other, creating a multilayered capsule (Strand and Pech, 1995). Lamellocytes are particularly involved in encapsulating and neutralizing invading pathogens that are too large to be up-taken by phagocytosis (Lemaitre and Hoffmann, 2007). The third stage of encapsulation involves crystal cells that account for 5% of the larval hemocytes. These cells function as storage sites for pro-phenoloxidase (PPO) and are therefore involved in the melanotic defensive response. Lysis of crystal cells triggers the melanization of the capsule surface (Strand and Pech, 1995; Fellowes and Godfray, 2000; Lemaitre and Hoffmann, 2007). Within the capsule, the encapsulated parasitoid egg gets killed by either direct asphyxiation (Salt, 1970) or by the release of superoxide anions or hydroxyl radicals from the capsule content (Nappi et al., 1995, 2000; Nappi and Vass, 1998) (Figure 3).
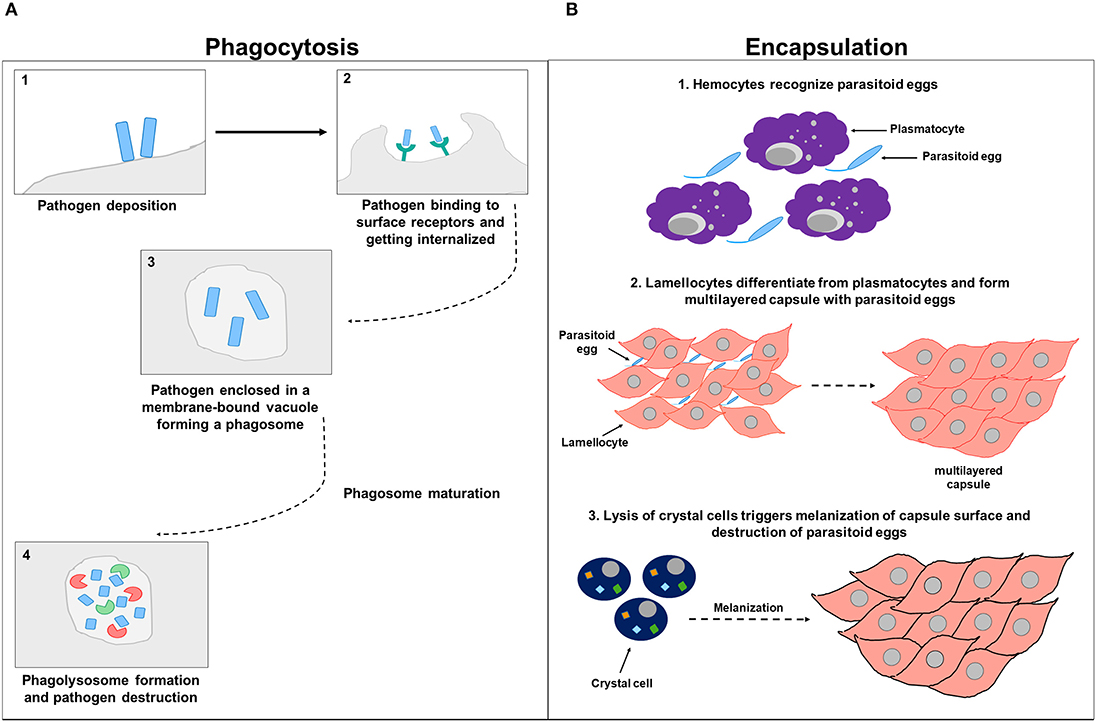
Figure 3. Cell-mediated immunity. (A) Represents a schematic diagram of phagocytosis. After the pathogen deposits on the host cell surface (1), it binds to phagocytic receptors and gets internalized (2) and enclosed in a membrane-bound vacuole forming a phagosome (3). The phagosome undergoes subsequent phases of maturation before eventually forming a phagolysosome that contains factors including DNases and proteases involved in pathogen destruction (4). (B) Represents a schematic diagram of encapsulation. In the first stage of encapsulation, hemocytes recognize parasitoid eggs as foreign invaders, triggering downstream signaling (1). In the second stage of encapsulation, hemocytes increase in numbers and lamellocytes differentiated from plasmatocytes and attach to parasitoid eggs and to each other, forming a multilayered capsule (2). In the third stage of encapsulation, crystal cells are involved and synthesize enzymes needed for melanization. Parasitoid eggs get sheathed, immobilized by the deposited melanin, and destroyed within the capsule either by direct asphyxiation or by the release of superoxide anions or hydroxyl radicals (3).
Melanization is another prominent immune response in insects characterized by melanin synthesis and deposition around intruding microorganisms (Christensen et al., 2005). Melanization is also involved in wound healing, phagocytosis, blood coagulation, and AMP expression in arthropods (Ashida and Brey, 1995; Söderhäll and Cerenius, 1998; Cerenius et al., 2008). The melanotic reaction, which is generally induced by either a pathogenic infection or tissue injury, culminates in the proteolytic cleavage of inactive PPO to active phenol oxidase (PO), the chief enzyme in melanin biogenesis (Cerenius et al., 2008). To avoid the production of excessive intermediates that are toxic to the host, the activation of melanization is normally tightly regulated (De Gregorio et al., 2002b; Ligoxygakis et al., 2002b; Scherfer et al., 2008; Tang et al., 2008). In Drosophila, genetic studies identified melanization regulators including serine proteases and serpin proteins. MP1 and MP2/sp7/PAE1 clip proteases, for example, positively regulate melanization. Silencing either MP1 or MP2 inhibit PO activation upon pathogenic infection (De Gregorio et al., 2002b; Ligoxygakis et al., 2002b; Castillejo-Lopez and Hacker, 2005; Scherfer et al., 2008; Tang et al., 2008). Several studies also highlighted a role of PGRPs in inducing melanization. The proPO cascade in Drosophila larvae is induced by a forced expression of PGRP-LE, independent of infection. Consistent with this, PGRP-LE is required for infection (E. coli)-induced melanization (Takehana et al., 2002, 2004). Likewise, PGRP-LC regulates melanization in Drosophila (Schmidt et al., 2007). Although melanization is considered an integral component in insect immunity, evidence of direct killing through quinone synthesis and melanin production has been reported in a few studies in insect species only. In Anopheles gamabiae, melanization was shown to retard Beauveria bassiana growth and dissemination (Yassine et al., 2012). In Manduca sexta, however, 60–94% killing of a broad spectrum of gram-negative bacterial species including Klebsiella pneumoniae, Escherichia coli, Pseudomonas aeruginosa, and Salmonella typhimurium, and 52–99% killing of gram-positive bacterial species including Staphylococcus aureus, Bacillus subtilis, Bacillus cereus, and Micrococcus luteus was reported in an active melanotic milieu (Zhao et al., 2007). Similarly, a recent study in D. melanogaster publicitized a novel role of melanization in anti-nematode immunity (Cooper et al., 2019). It is worth noting here that some host–pathogen interactions are “genotype by genotype” driven. Drosophila melanogaster's melanotic and complement-like immunity, for example, vary extensively against the parasitoid wasp Leptopilina boulardi. PO activity is predominantly affected by the host genotype, while TEP1 upregulation is controlled by the parasite genotype itself. Lamellocyte differentiation, on the other hand, depends on the specific combination of both the host and parasite genotypes (Leitão et al., 2019).
RNA Interference
The RNA interference (RNAi) pathway, which suppresses gene expression through targeted RNA degradation, embodies an ancient mechanism of anti-viral immunity in plants, nematodes, and arthropods including Drosophila (Hamilton and Baulcombe, 1999; Li et al., 2002; Lu et al., 2005; Wilkins et al., 2005; Cherry and Silverman, 2006; Wang et al., 2006; Zambon et al., 2006; Saleh et al., 2009; Karlikow et al., 2014). This pathway emerges in two major phases including the “initiation” and the “execution” phase. Either endogenous (short hairpin RNAs manufactured by the genome, perversely expressed trans-genes, and transposons) or exogenous sources (naturally occurring or experimentally made dsRNA) can introduce dsRNA to initiate RNAi (Hannon, 2002; Zambon et al., 2006). dsRNA are recognized and cleaved by Dicer molecules to form small RNAs (Hammond et al., 2000; Blaszczyk et al., 2001; Zambon et al., 2006) that get integrated into the RNA-induced silencing complex (RISC), denoting the execution phase of the RNAi pathway (Blaszczyk et al., 2001; Nykanen et al., 2001; Zambon et al., 2006). Unlike mammals that have only one Dicer gene, and which is difficult to study, flies possess two genes, Dicer1 and Dicer2, that are required for processing miRNA precursors from pre-miRNA and siRNA precursors from long dsRNA, respectively (Robles-Sikisaka et al., 2001). The single strand of either miRNA or siRNA integrated into the RISC complex acts as a platform for RISC to recognize complementary messenger RNA (mRNA) transcript. Upon recognition, Argonaute, one of the proteins in RISC, activates and cleaves the mRNA, inhibiting antiviral functions and suppressing viral expression (Karlikow et al., 2014) (Figure 4). Other existing, yet poorly identified, RNAi pathways include the PIWI-interacting RNA (piRNA) pathway that shields host cells from endogenous mobile genetic elements (Buchon et al., 2014). Several studies in Drosophila reported that loss-of-function mutations in essential RNAi pathway genes increase host vulnerability to viral infection (Zambon et al., 2006; Aliyari et al., 2008; Buchon et al., 2014). In mammals, other antiviral defense strategies, including protein sensors that recognize viral dsRNA motifs, have been identified. Among these sensors are the DEAD-box helicases RIG-I (Retinoic acid-inducible gene I) and MDA5 (Melanoma Differentiation-Associated protein 5), together known as RIG-I-like receptors (RLRs). Upon recognizing viral nucleic acid during the primary viral infection stages, these sensors induce the expression of type 1 interferons (IFNα and IFNβ) and other pro-inflammatory cytokines (Song and Rossi, 2017; van der Veen et al., 2018; Brisse and Ly, 2019). Interestingly, studies in Drosophila showed that Dicer-2 closely resembles the mammalian RLRs, not only by cleaving dsRNA into siRNA, but also by activating the transcription of antiviral effectors proteins (Deddouche et al., 2008). Genetic screening in the fruit fly revealed additional antiviral roles of DEAD-box helicase. DDX17 (known as Rm62), for example, exhibits antiviral activity against arthropod-borne bunyaviruses (Deddouche et al., 2008). In addition to these nucleic acid-elicited responses, some viruses can be directly recognized by Toll-7, which promotes the activation of antiviral autophagy in an AKT pathway-dependent manner through phosphoinositide 3-kinase (PI3K) and target of rapamycin (Tor) (Buchon et al., 2014). Likewise, other studies in Drosophila also revealed a direct antiviral role of autophagy against the vesicular stomatitis virus, initiated by the pathogen surface glycoprotein VSVG (Shelly et al., 2009). Sabin et al. identified Ars2 (CG7843) as a key element of Drosophila antiviral immunity using an RNAi library and demonstrated that a loss of Ars2 function in either cells or flies promotes vulnerability to RNA viruses. In addition to its antiviral characteristic, Ars2 was shown to modulate Dcr-2 activity in vitro by physically interacting with it. It was also shown to play an essential role in siRNA- and miRNA-mediated silencing. This crucial role of Ars2 in these small RNA pathways delivers novel insight into the biogenesis of small RNAs, a platform that could be extended to other systems (Sabin et al., 2009). Similarly, unrecognized host genes imperative for the influenza viral replication have been identified using genome-wide RNAi screens in Drosophila. Three of these identified genes have corresponding homologs in humans (ATP6V0D1, COX6A1, and NXF1). When tested in human HEK 293 cells, these genes were shown to be involved in the replication of H5N1 and H1N1 influenza A viruses, but not in vaccinia nor in vesicular stomatitis viral replication (Hao et al., 2008). The natural resistance-associated macrophage (NRAMP), a divalent metal ion transporter and a cell surface molecule expressed on Drosophila cells and required for binding and entry of sindbis virus to host cells, was also identified using RNAi technology. dNRAMP mutant flies were shown to be protected from viral infection (Rose et al., 2011). Interestingly, Carpenter et al. identified many differentially expressed genes in sigma virus-infected flies, several of which are neither up-regulated by bacterial or fungal infection, nor controlled by Toll, IMD, or JAK/STAT pathways, implying the involvement of other distinct regulatory immune mechanisms in defense against sigma virus in infected flies (Carpenter et al., 2009).
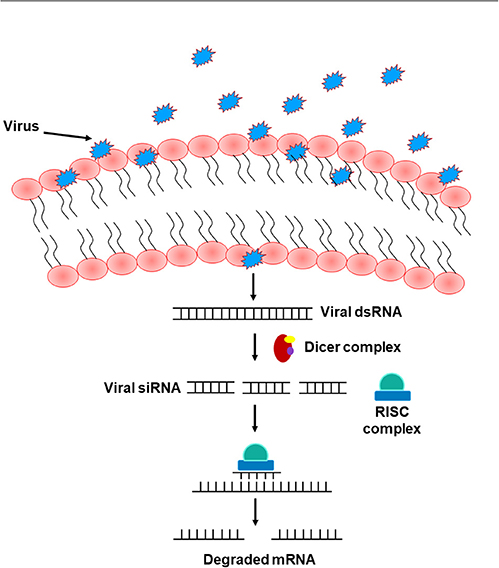
Figure 4. The RNA interference pathway. Upon entry into the cells, viruses shed their shielding external coat, uncovering their RNA, and forming dsRNA. This formed dsRNA gets recognized by the Dicer complex and processed to form viral siRNA. A single strand of this siRNA gets incorporated into the RISC complex and act as a template to recognize complementary mRNA, resulting in mRNA cleavage and therefore silencing of viral RNA.
Ongoing studies are now applying genome-wide association study (GWAS) to identify the genetic basis of natural variation in Drosophila's immunity against pathogens. A study by Chapman et al. identified single nucleotide polymorphisms associated with genes (Bomanin gene BomBc1, krishah, and S6k) that significantly affected the fly's immunity to Enterococcus faecalis infection. Surprisingly, none of these genes are classified as canonical immune genes (Chapman et al., 2020).
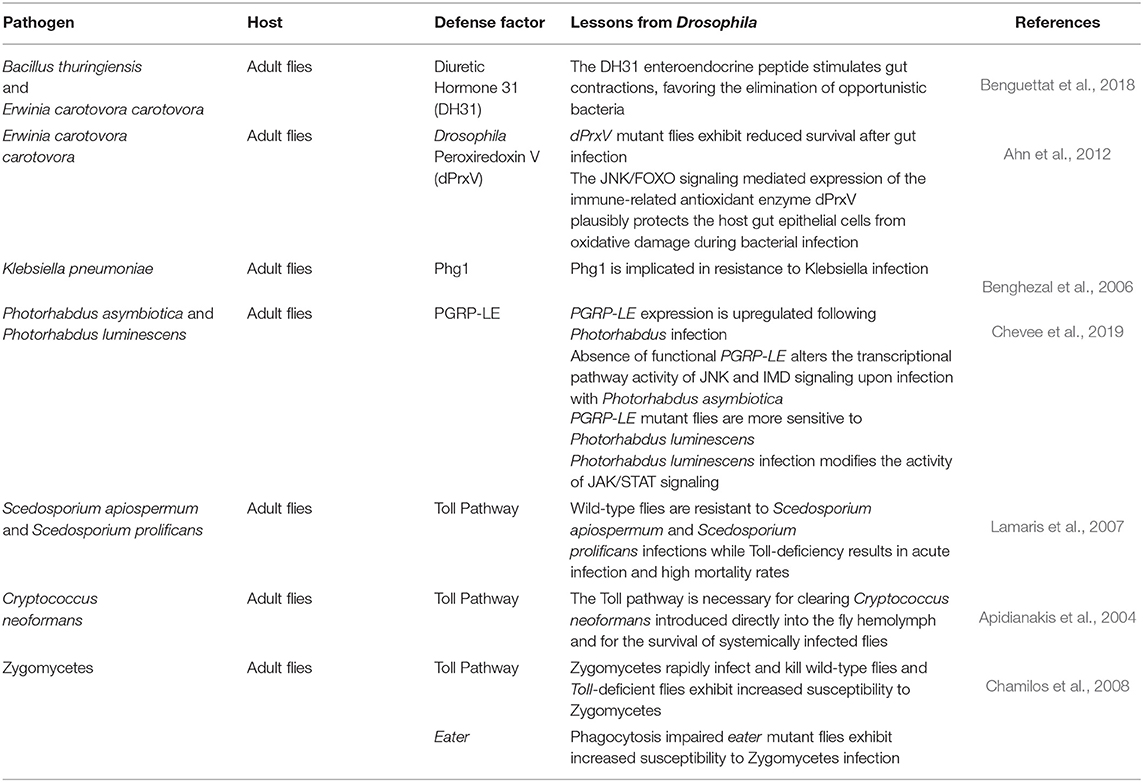
Currently, the direction in unraveling host defense factors and innate immune effector molecules in the Drosophila model organism is heading toward bracketing classical genetic approaches with GWAS and genome-wide RNAi screening of flies with either loss of function or over-expressed immune genes, in addition to the use of co-immunoprecipitation assays and mass spectrometry to identify immune protein complexes. Moreover, bioinformatics analysis is being extensively adapted in deciphering candidate molecules and post-translational alterations that could impact the host's immune signaling pathways (Kanoh et al., 2019; Chapman et al., 2020). Nevertheless, and along with the ongoing high-throughput screens to discover conserved genes involved in host–pathogen interactions and immune signaling, the CRISPR/Cas9 technology has paved the way for additional wide-scale-based screens in Drosophila cultured cells, the results of which could be followed up in vivo in flies and/or mammals (Viswanatha et al., 2019). Some identified host factors required for defense against a broad spectrum of pathogens are summarized in Table 1.
Pathogen Virulence Factors
Diverse well-designed screening assay systems have been established to identify virulence factors contributing to pathogen-induced host killing using the Drosophila model organism. Screening for virulence-attenuated mutants identified a set of genes involved in the multi-host pathogenesis of P. aeruginosa PA14, for example. Follow-up studies further characterized these genes to validate the use of Drosophila as a model for high-throughput identification of novel virulence factors. Characterizing hudR, an identified virulence gene encoding a MarR/SlyA family transcription factor, for instance, revealed that eminent expression of hudA (homologous to UbiD) is required and adequate to attenuate the virulence of hudR mutants in infected flies (Kim et al., 2008). Since quorum sensing is involved in the pathogenicity of P. aeruginosa, several studies focused on identifying quorum sensing-regulated virulence factors, as an appealing therapeutic approach to control P. aeruginosa infection (Bjarnsholt and Givskov, 2007). P. aeruginosa oxylipin lipids were also identified as pathogenic factors that promote bacterial virulence and biofilm formation in Drosophila (Martinez and Campos-Gomez, 2016). Further studies in P. aeruginosa also revealed that the phosphorylation state of the transcriptional response regulator AlgR inversely controls the production of pyoverdine and pyocyanin, two important P. aeruginosa virulence factors (Little et al., 2018). Other studies also focused on deciphering the effect of bacterial toxins on the host using fruit flies. P. aeruginosa exotoxin ExoS was shown to affect the activity of Rho GTPases, as the directed expression of the bacterial ExoS GAP domain (ExoSGAP) inhibits Rac1-, Cdc42-, and Rho-dependent signaling, suppressing Drosophila cellular immunity (Avet-Rochex et al., 2005). V. cholerae toxin, in turn, was shown to decrease intestinal stem cell division, alter epithelial regeneration, and induce cell–cell junctional damage (Guichard et al., 2013; Wang et al., 2013). Interestingly, the Vibrio polysaccharide (VPS)-dependent biofilm, which is highly activated upon entry into the arthropod intestine, is essential for Drosophila intestinal colonization (Purdy and Watnick, 2011). Surprisingly however, quorum sensing promotes a more auspicious interaction between the fly host and V. cholerae by reducing the nutritional burden of intestinal colonization in the host (Kamareddine et al., 2018b). Novel H. pylori effector proteins like the cytotoxin-associated gene A (CagA) have been studied in transgenic Drosophila flies. CagA mimics the eukaryotic adaptor protein Grb2-associated binder (Gab) and activates phosphatase SHP-2, a component of the receptor tyrosine kinase pathways. These findings in D. melanogaster could provide more insight into the role of translocated bacterial proteins that targets highly conserved eukaryotic cellular processes (Botham et al., 2008). The Anthrax toxin produced by Bacillus anthracis is comprised of protective antigen (PA), edema factor (EF), and lethal factor (LF) (Lacy and Collier, 2002). Similar to their function in mammals, LF cleaves MAPK kinases, and EF inhibits hedgehog pathway in flies (Guichard et al., 2006). This similarity in function strengthens the argument of choosing Drosophila as a multicellular host system to study in vivo function of virulence factors and diverse toxins. Drosophila has been also extensively used to study infectious properties of several bacterial species like Porphyromonas gingivalis (W83), a gram-negative obligate anaerobic bacteria strongly implicated in adult periodontitis (Griffen et al., 1998; Ezzo and Cutler, 2003; Igboin et al., 2011). P. gingivalis causes systemic infection in Drosophila and promotes potent killing in a dose-dependent manner. Interestingly, both heat-killed and live P. gingivalis are similarly pathogenic to the fly, suggesting a role of P. gingivalis cell surface components and Drosophila immunity in dictating pathology in this host–pathogen model (Igboin et al., 2011). Tabuchi et al. demonstrated an important role of dltA, a gene responsible for D-alanylation of techoic acid in the cell wall of gram-positive S. aureus, in inhibiting the fly's Toll pathway. S. aureus-infected dltA mutant flies exhibited an increase in life span compared to flies expressing dltA normally (Tabuchi et al., 2010). Fungal virulence factors have been also reported in several Drosophila studies. Gliotoxin, for example, contributes to the virulence of Aspergillus fumigatus in fruit flies with functional phagocytes as well as in non-neutropenic mice, suggesting that gliotoxin principally targets neutrophils or other phagocytes (Spikes et al., 2008). Cas5 transcription in Candida albicans regulates cell wall integrity and is essential for fungal virulence in both murine and Toll mutant flies (Chamilos et al., 2009). Likewise, Candida glabrata mutant strains lacking the yapsin virulence factors or the high-osmolarity glycerol pathway exhibit a less virulent effect in infected flies (Quintin et al., 2013). Several antifungal drug efficacy studies against invasive aspergillosis (Lionakis et al., 2005) and malasseziosis (Merkel et al., 2018) have been conducted in the Drosophila model. Many studies have also demonstrated the ability of parasitic nematodes, like those belonging to the Heterorhabditis genus, to infect and kill fruit flies at larval and adult stages and to trigger an up-regulation in several genes belonging to the Toll, IMD, JAK/STAT, and TGF-β signaling pathways (Castillo et al., 2013, 2015; Arefin et al., 2014). Heterorhabditis gerrardi, for example, harbors the pathogenic Photorhabdus asymbiotica bacteria, which gets ejected from the nematode gut into the host's hemolymph. In the hemolymph, Photorhabdus asymbiotica proliferates and releases toxins and virulence factors that kills the host and provides a favorable environment for the nematode (Waterfield et al., 2008; Eleftherianos et al., 2010). To analyze the impact of the Photorhabdus-Heterorhabditis mutualistic relation on the transcriptional profiles of the host, Castillo et al. (2015) performed next-generation RNA-sequencing on flies infected with Photorhabdus alone, germ-free Heterorhabditis lacking Photorhabdus, and Heterorhabditis carrying Photorhabdus. The bioinformatics analysis of that study revealed an impact of Photorhabdus on the transcription of fly genes associated with translational repression and stress responses, and an effect of Heterorhabditis on the expression profiles of genes involved in metabolism, lipid homeostasis, stress responses, DNA/protein synthesis, and functions of the nervous system.
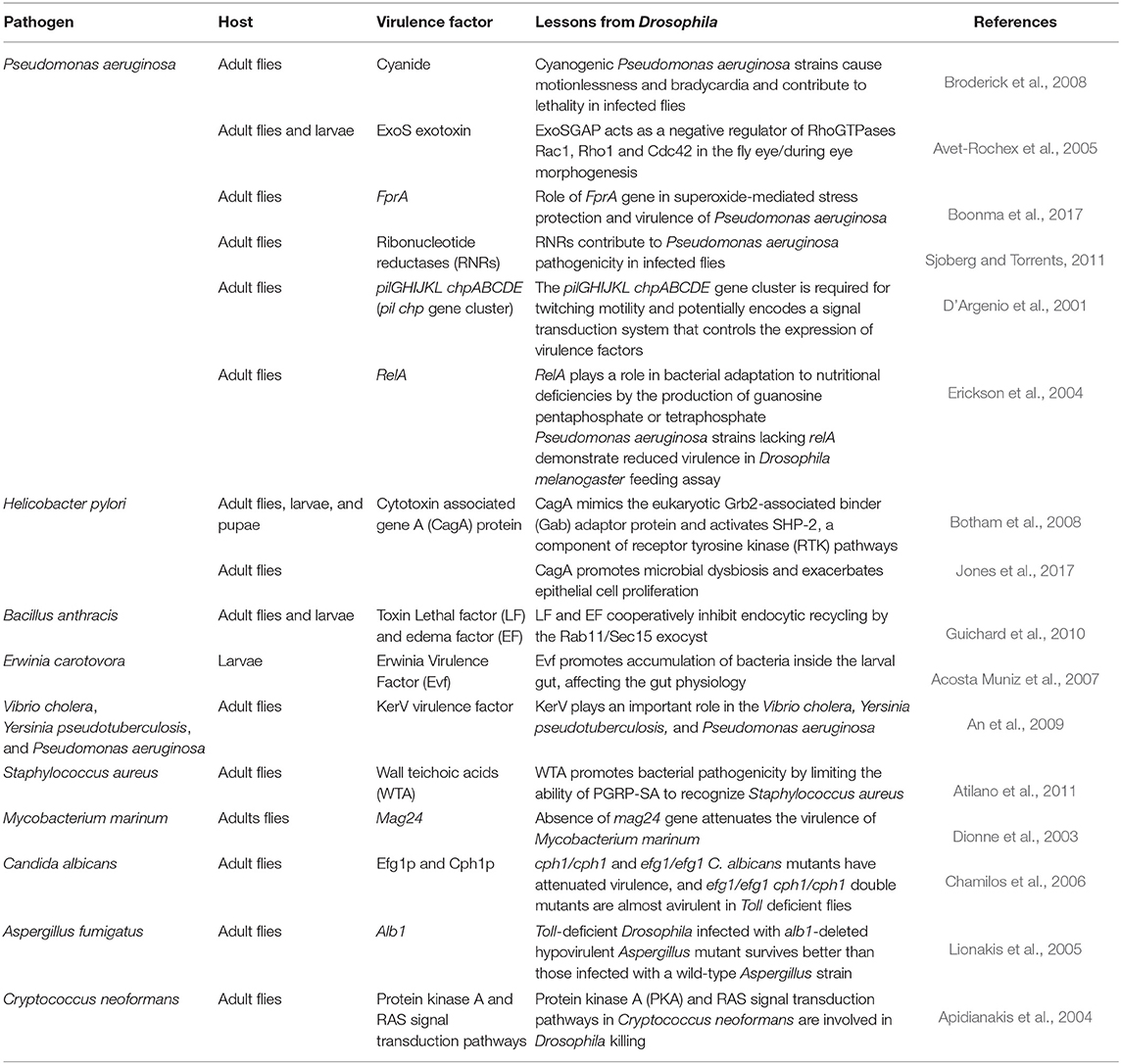
In the last few years, a number of studies have examined virulence factors of human viral pathogens in Drosophila, as the fruit fly model facilitates the implementation of systematic, genome-wide RNAi analysis commonly used to identify genes that are involved in viral replication (Kuttenkeuler and Boutros, 2004). Since a broad spectrum of RNA viruses exploit internal ribosome entry sites (IRESs) for translation, genome-wide RNAi screen in Drosophila cells infected with Drosophila C virus were performed to reveal host factors required for IRES-dependent translation and viral replication. A study by Cherry et al. revealed 66 ribosomal proteins needed for Drosophila C virus, but not for non-IRES-containing RNA virus (Cherry et al., 2005). Some identified pathogen virulence factors are summarized in Table 2.
Conclusion
The use of animal models serves as a foundation to reveal conserved aspects of human disease. Unraveling detailed mechanisms of host–pathogen interactions using Drosophila provides further insight into the pathogenic arm of a microorganism and the defensive arm of a host. A better understanding of host–microbe interactions is desirable for the development of novel successful treatment regimens for pathogen-caused diseases.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Qatar University (QUST-1-CHS-2020-4) for funding this publication.
References
Acosta Muniz, C., Jaillard, D., Lemaitre, B., and Boccard, F. (2007). Erwinia carotovora Evf antagonizes the elimination of bacteria in the gut of Drosophila larvae. Cell Microbiol. 9, 106–119. doi: 10.1111/j.1462-5822.2006.00771.x
Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195. doi: 10.1126/science.287.5461.2185
Agaisse, H., Burrack, L. S., Philips, J. A., Rubin, E. J., Perrimon, N., and Higgins, D. E. (2005). Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science 309, 1248–1251. doi: 10.1126/science.1116008
Agaisse, H., and Perrimon, N. (2004). The roles of JAK/STAT signaling in Drosophila immune responses. Immunol. Rev. 198, 72–82. doi: 10.1111/j.0105-2896.2004.0133.x
Ahn, H. M., Lee, K. S., Lee, D. S., and Yu, K. (2012). JNK/FOXO mediated PeroxiredoxinV expression regulates redox homeostasis during Drosophila melanogaster gut infection. Dev. Comp. Immunol. 38, 466–473. doi: 10.1016/j.dci.2012.07.002
Akira, S., Uematsu, S., and Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cell 124, 783–801. doi: 10.1016/j.cell.2006.02.015
Aliyari, R., Wu, Q., Li, H. W., Wang, X. H., Li, F., Green, L. D., et al. (2008). Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe 4, 387–397. doi: 10.1016/j.chom.2008.09.001
An, D., Apidianakis, Y., Boechat, A. L., Baldini, R. L., Goumnerov, B. C., and Rahme, L. G. (2009). The pathogenic properties of a novel and conserved gene product, KerV, in proteobacteria. PLoS ONE 4:e7167. doi: 10.1371/journal.pone.0007167
Anjum, S. G., Xu, W., Nikkholgh, N., Basu, S., Nie, Y., Thomas, M., et al. (2013). Regulation of Toll signaling and inflammation by β-arrestin and the SUMO protease Ulp1. Genetics 195, 1307–1317. doi: 10.1534/genetics.113.157859
Apidianakis, Y., Rahme, L. G., Heitman, J., Ausubel, F. M., Calderwood, S. B., and Mylonakis, E. (2004). Challenge of Drosophila melanogaster with Cryptococcus neoformans and role of the innate immune response. Eukaryot. Cell 3, 413–419. doi: 10.1128/EC.3.2.413-419.2004
Arefin, B., Kucerova, L., Dobes, P., Markus, R., Strnad, H., Wang, Z., et al. (2014). Genome-wide transcriptional analysis of Drosophila larvae infected by entomopathogenic nematodes shows involvement of complement, recognition and extracellular matrix proteins. J. Innate Immun. 6, 192–204. doi: 10.1159/000353734
Ashida, M., and Brey, P. T. (1995). Role of the integument in insect defense: pro-phenol oxidase cascade in the cuticular matrix. Proc. Natl. Acad. Sci. U.S.A. 92, 10698–10702. doi: 10.1073/pnas.92.23.10698
Atilano, M. L., Yates, J., Glittenberg, M., Filipe, S. R., and Ligoxygakis, P. (2011). Wall teichoic acids of Staphylococcus aureus limit recognition by the drosophila peptidoglycan recognition protein-SA to promote pathogenicity. PLoS Pathog. 7:e1002421. doi: 10.1371/journal.ppat.1002421
Avadhanula, V., Weasner, B. P., Hardy, G. G., Kumar, J. P., and Hardy, R. W. (2009). A novel system for the launch of alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. PLoS Pathog. 5:e1000582. doi: 10.1371/journal.ppat.1000582
Avet-Rochex, A., Bergeret, E., Attree, I., Meister, M., and Fauvarque, M. O. (2005). Suppression of Drosophila cellular immunity by directed expression of the ExoS toxin GAP domain of Pseudomonas aeruginosa. Cell Microbiol. 7, 799–810. doi: 10.1111/j.1462-5822.2005.00512.x
Aymeric, J. L., Givaudan, A., and Duvic, B. (2010). Imd pathway is involved in the interaction of Drosophila melanogaster with the entomopathogenic bacteria, Xenorhabdus nematophila and Photorhabdus luminescens. Mol. Immunol. 47, 2342–2348. doi: 10.1016/j.molimm.2010.05.012
Benghezal, M., Fauvarque, M. O., Tournebize, R., Froquet, R., Marchetti, A., Bergeret, E., et al. (2006). Specific host genes required for the killing of Klebsiella bacteria by phagocytes. Cell Microbiol. 8, 139–148. doi: 10.1111/j.1462-5822.2005.00607.x
Benguettat, O., Jneid, R., Soltys, J., Loudhaief, R., Brun-Barale, A., Osman, D., et al. (2018). The DH31/CGRP enteroendocrine peptide triggers intestinal contractions favoring the elimination of opportunistic bacteria. PLoS Pathog. 14:e1007279. doi: 10.1371/journal.ppat.1007279
Bier, E., and Guichard, A. (2012). Deconstructing host-pathogen interactions in Drosophila. Dis. Models Mech. 5, 48–61. doi: 10.1242/dmm.000406
Bjarnsholt, T., and Givskov, M. (2007). The role of quorum sensing in the pathogenicity of the cunning aggressor Pseudomonas aeruginosa. Anal. Bioanal. Chem. 387, 409–414. doi: 10.1007/s00216-006-0774-x
Blandin, S., Shiao, S.-H., Moita, L. F., Janse, C. J., Waters, A. P., Kafatos, F. C., et al. (2004). Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell 116, 661–670. doi: 10.1016/S0092-8674(04)00173-4
Blaszczyk, J., Tropea, J. E., Bubunenko, M., Routzahn, K. M., Waugh, D. S., Court, D. L., et al. (2001). Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure 9, 1225–1236. doi: 10.1016/S0969-2126(01)00685-2
Boonma, S., Romsang, A., Duang-Nkern, J., Atichartpongkul, S., Trinachartvanit, W., Vattanaviboon, P., et al. (2017). The FinR-regulated essential gene fprA, encoding ferredoxin NADP+ reductase: roles in superoxide-mediated stress protection and virulence of Pseudomonas aeruginosa. PLoS ONE 12:e0172071. doi: 10.1371/journal.pone.0172071
Botham, C. M., Wandler, A. M., and Guillemin, K. (2008). A transgenic Drosophila model demonstrates that the Helicobacter pylori CagA protein functions as a eukaryotic Gab adaptor. PLoS Pathog. 4:e1000064. doi: 10.1371/journal.ppat.1000064
Brand, A. H., and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415.
Bretscher, A. J., Honti, V., Binggeli, O., Burri, O., Poidevin, M., Kurucz, E., et al. (2015). The Nimrod transmembrane receptor Eater is required for hemocyte attachment to the sessile compartment in Drosophila melanogaster. Biol. Open 4, 355–363. doi: 10.1242/bio.201410595
Brisse, M., and Ly, H. (2019). Comparative structure and function analysis of the RIG-I-like receptors: RIG-I and MDA5. Front. Immunol. 10:1586. doi: 10.3389/fimmu.2019.01586
Broderick, K. E., Chan, A., Balasubramanian, M., Feala, J., Reed, S. L., Panda, M., et al. (2008). Cyanide produced by human isolates of Pseudomonas aeruginosa contributes to lethality in Drosophila melanogaster. J. Infect. Dis. 197, 457–464. doi: 10.1086/525282
Buchon, N., Broderick, N. A., Chakrabarti, S., and Lemaitre, B. (2009b). Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 23, 2333–2344. doi: 10.1101/gad.1827009
Buchon, N., Broderick, N. A., Poidevin, M., Pradervand, S., and Lemaitre, B. (2009c). Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe 5, 200–211. doi: 10.1016/j.chom.2009.01.003
Buchon, N., Poidevin, M., Kwon, H. M., Guillou, A., Sottas, V., Lee, B. L., et al. (2009a). A single modular serine protease integrates signals from pattern-recognition receptors upstream of the Drosophila Toll pathway. Proc. Natl. Acad. Sci. U.S.A. 106, 12442–12447. doi: 10.1073/pnas.0901924106
Buchon, N., Silverman, N., and Cherry, S. (2014). Immunity in Drosophila melanogaster–from microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 14, 796–810. doi: 10.1038/nri3763
Carpenter, J., Hutter, S., Baines, J. F., Roller, J., Saminadin-Peter, S. S., Parsch, J., et al. (2009). The transcriptional response of Drosophila melanogaster to infection with the sigma virus (Rhabdoviridae). PLoS ONE 4:e6838. doi: 10.1371/journal.pone.0006838
Castillejo-Lopez, C., and Hacker, U. (2005). The serine protease Sp7 is expressed in blood cells and regulates the melanization reaction in Drosophila. Biochem. Biophys. Res. Commun. 338, 1075–1082. doi: 10.1016/j.bbrc.2005.10.042
Castillo, J. C., Creasy, T., Kumari, P., Shetty, A., Shokal, U., Tallon, L. J., et al. (2015). Drosophila anti-nematode and antibacterial immune regulators revealed by RNA-Seq. BMC Genomics 16:519. doi: 10.1186/s12864-015-1690-2
Castillo, J. C., Shokal, U., and Eleftherianos, I. (2013). Immune gene transcription in Drosophila adult flies infected by entomopathogenic nematodes and their mutualistic bacteria. J. Insect. Physiol. 59, 179–185. doi: 10.1016/j.jinsphys.2012.08.003
Cerenius, L., Lee, B. L., and Söderhäll, K. (2008). The proPO-system: pros and cons for its role in invertebrate immunity. Trends Immunol. 29, 263–271. doi: 10.1016/j.it.2008.02.009
Chamilos, G., Bignell, E. M., Schrettl, M., Lewis, R. E., Leventakos, K., May, G. S., et al. (2010). Exploring the concordance of Aspergillus fumigatus pathogenicity in mice and Toll-deficient flies. Med. Mycol. 48, 506–510. doi: 10.3109/13693780903225813
Chamilos, G., Lewis, R. E., Hu, J., Xiao, L., Zal, T., Gilliet, M., et al. (2008). Drosophila melanogaster as a model host to dissect the immunopathogenesis of zygomycosis. Proc. Natl. Acad. Sci. U.S.A. 105, 9367–9372. doi: 10.1073/pnas.0709578105
Chamilos, G., Lionakis, M. S., Lewis, R. E., Lopez-Ribot, J. L., Saville, S. P., Albert, N. D., et al. (2006). Drosophila melanogaster as a facile model for large-scale studies of virulence mechanisms and antifungal drug efficacy in Candida species. J. Infect. Dis. 193, 1014–1022. doi: 10.1086/500950
Chamilos, G., Nobile, C. J., Bruno, V. M., Lewis, R. E., Mitchell, A. P., and Kontoyiannis, D. P. (2009). Candida albicans Cas5, a regulator of cell wall integrity, is required for virulence in murine and toll mutant fly models. J. Infect. Dis. 200, 152–157. doi: 10.1086/599363
Chapman, J. R., Dowell, M. A., Chan, R., and Unckless, R. L. (2020). The genetic basis of natural variation in Drosophila melanogaster immune defense against Enterococcus faecalis. Genes 11:234. doi: 10.3390/genes11020234
Cherry, S. (2008). Genomic RNAi screening in Drosophila S2 cells: what have we learned about host-pathogen interactions? Curr. Opin. Microbiol. 11, 262–270. doi: 10.1016/j.mib.2008.05.007
Cherry, S., Doukas, T., Armknecht, S., Whelan, S., Wang, H., Sarnow, P., et al. (2005). Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 19, 445–452. doi: 10.1101/gad.1267905
Cherry, S., and Silverman, N. (2006). Host-pathogen interactions in drosophila: new tricks from an old friend. Nat. Immunol. 7, 911–917. doi: 10.1038/ni1388
Chevee, V., Sachar, U., Yadav, S., Heryanto, C., and Eleftherianos, I. (2019). The peptidoglycan recognition protein PGRP-LE regulates the Drosophila immune response against the pathogen Photorhabdus. Microb. Pathog. 136:103664. doi: 10.1016/j.micpath.2019.103664
Christensen, B. M., Li, J., Chen, C.-C., and Nappi, A. J. (2005). Melanization immune responses in mosquito vectors. Trends Parasitol. 21, 192–199. doi: 10.1016/j.pt.2005.02.007
Cooper, D., Wuebbolt, C., Heryanto, C., and Eleftherianos, I. (2019). The prophenoloxidase system in Drosophila participates in the anti-nematode immune response. Mol. Immunol. 109, 88–98. doi: 10.1016/j.molimm.2019.03.008
Costa, A., Jan, E., Sarnow, P., and Schneider, D. (2009). The Imd pathway is involved in antiviral immune responses in Drosophila. PLoS ONE 4:e7436. doi: 10.1371/journal.pone.0007436
Cronin, S. J., Nehme, N. T., Limmer, S., Liegeois, S., Pospisilik, J. A., Schramek, D., et al. (2009). Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science 325, 340–343. doi: 10.1126/science.1173164
Cuttell, L., Vaughan, A., Silva, E., Escaron, C. J., Lavine, M., Van Goethem, E., et al. (2008). Undertaker, a Drosophila Junctophilin, links Draper-mediated phagocytosis and calcium homeostasis. Cell 135, 524–534. doi: 10.1016/j.cell.2008.08.033
D'Argenio, D. A., Gallagher, L. A., Berg, C. A., and Manoil, C. (2001). Drosophila as a model host for Pseudomonas aeruginosa infection. J. Bacteriol. 183, 1466–1471. doi: 10.1128/JB.183.4.1466-1471.2001
De Gregorio, E., Han, S.-J., Lee, W.-J., Baek, M.-J., Osaki, T., Kawabata, S.-I., et al. (2002b). An immune-responsive Serpin regulates the melanization cascade in Drosophila. Dev. Cell 3, 581–592. doi: 10.1016/S1534-5807(02)00267-8
De Gregorio, E., Spellman, P. T., Tzou, P., Rubin, G. M., and Lemaitre, B. (2002a). The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 21, 2568–2579. doi: 10.1093/emboj/21.11.2568
Deddouche, S., Matt, N., Budd, A., Mueller, S., Kemp, C., Galiana-Arnoux, D., et al. (2008). The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat. Immunol. 9, 1425–1432. doi: 10.1038/ni.1664
Dionne, M. S., Ghori, N., and Schneider, D. S. (2003). Drosophila melanogaster is a genetically tractable model host for Mycobacterium marinum. Infect. Immun. 71, 3540–3550. doi: 10.1128/IAI.71.6.3540-3550.2003
Dionne, M. S., and Schneider, D. S. (2008). Models of infectious diseases in the fruit fly Drosophila melanogaster. Dis. Models Mech. 1, 43–49. doi: 10.1242/dmm.000307
Dostert, C., Jouanguy, E., Irving, P., Troxler, L., Galiana-Arnoux, D., Hetru, C., et al. (2005). The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat. Immunol. 6, 946–953. doi: 10.1038/ni1237
Eleftherianos, I., ffrench-Constant, R.H., Clarke, D. J., Dowling, A. J., and Reynolds, S. E. (2010). Dissecting the immune response to the entomopathogen Photorhabdus. Trends Microbiol. 18, 552–560. doi: 10.1016/j.tim.2010.09.006
Engel, E., Viargues, P., Mortier, M., Taillebourg, E., Couté, Y., Thevenon, D., et al. (2014). Identifying USPs regulating immune signals in Drosophila: USP2 deubiquitinates Imd and promotes its degradation by interacting with the proteasome. Cell Commun. Signal. 12:41. doi: 10.1186/s12964-014-0041-2
Erickson, D. L., Lines, J. L., Pesci, E. C., Venturi, V., and Storey, D. G. (2004). Pseudomonas aeruginosa relA contributes to virulence in Drosophila melanogaster. Infect. Immun. 72, 5638–5645. doi: 10.1128/IAI.72.10.5638-5645.2004
Ezzo, P. J., and Cutler, C. W. (2003). Microorganisms as risk indicators for periodontal disease. Periodontology (2000) 32, 24–35. doi: 10.1046/j.0906-6713.2003.03203.x
Fellowes, M. D., and Godfray, H. C. (2000). The evolutionary ecology of resistance to parasitoids by Drosophila. Heredity (Edinb). 84, 1–8. doi: 10.1046/j.1365-2540.2000.00685.x
Franc, N. C., Heitzler, P., Ezekowitz, R. A., and White, K. (1999). Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila. Science 284, 1991–1994. doi: 10.1126/science.284.5422.1991
Fullaondo, A., Garcia-Sanchez, S., Sanz-Parra, A., Recio, E., Lee, S. Y., and Gubb, D. (2011). Spn1 regulates the GNBP3-dependent Toll signaling pathway in Drosophila melanogaster. Mol. Cell Biol. 31, 2960–2972. doi: 10.1128/MCB.01397-10
Galiana-Arnoux, D., Dostert, C., Schneemann, A., Hoffmann, J. A., and Imler, J.-L. (2006). Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat. Immunol. 7, 590–597. doi: 10.1038/ni1335
Garver, L. S., Wu, J., and Wu, L. P. (2006). The peptidoglycan recognition protein PGRP-SC1a is essential for Toll signaling and phagocytosis of Staphylococcus aureus in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 103, 660–665. doi: 10.1073/pnas.0506182103
Gottar, M., Gobert, V., Matskevich, A. A., Reichhart, J.-M., Wang, C., Butt, T. M., et al. (2006). Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell 127, 1425–1437. doi: 10.1016/j.cell.2006.10.046
Govind, S. (2008). Innate immunity in Drosophila: pathogens and pathways. Insect Sci. 15, 29–43. doi: 10.1111/j.1744-7917.2008.00185.x
Griffen, A. L., Becker, M. R., Lyons, S. R., Moeschberger, M. L., and Leys, E. J. (1998). Prevalence of Porphyromonas gingivalis and periodontal health status. J. Clin. Microbiol. 36, 3239–3242. doi: 10.1128/JCM.36.11.3239-3242.1998
Guichard, A., Cruz-Moreno, B., Aguilar, B., van Sorge, N. M., Kuang, J., Kurkciyan, A. A., et al. (2013). Cholera toxin disrupts barrier function by inhibiting exocyst-mediated trafficking of host proteins to intestinal cell junctions. Cell Host Microbe 14, 294–305. doi: 10.1016/j.chom.2013.09.003
Guichard, A., McGillivray, S. M., Cruz-Moreno, B., van Sorge, N. M., Nizet, V., and Bier, E. (2010). Anthrax toxins cooperatively inhibit endocytic recycling by the Rab11/Sec15 exocyst. Nature 467, 854–858. doi: 10.1038/nature09446
Guichard, A., Park, J. M., Cruz-Moreno, B., Karin, M., and Bier, E. (2006). Anthrax lethal factor and edema factor act on conserved targets in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 103, 3244–3249. doi: 10.1073/pnas.0510748103
Haasnoot, J., and Berkhout, B. (2006). “RNA interference: its use as antiviral therapy,” in RNA Towards Medicine, eds V. Erdmann, J. Barciszewski, and J. Brosius (Berlin: Springer), 117–150. doi: 10.1007/3-540-27262-3_7
Hamilton, A. J., and Baulcombe, D. C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. doi: 10.1126/science.286.5441.950
Hammond, S. M., Bernstein, E., Beach, D., and Hannon, G. J. (2000). An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404, 293–296. doi: 10.1038/35005107
Hao, L., Sakurai, A., Watanabe, T., Sorensen, E., Nidom, C. A., Newton, M. A., et al. (2008). Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454, 890–893. doi: 10.1038/nature07151
Hedengren-Olcott, M., Olcott, M. C., Mooney, D. T., Ekengren, S., Geller, B. L., and Taylor, B. J. (2004). Differential activation of the NF-kappaB-like factors Relish and Dif in Drosophila melanogaster by fungi and Gram-positive bacteria. J. Biol. Chem. 279, 21121–21127. doi: 10.1074/jbc.M313856200
Herbein, G., and O'Brien, W. A. (2000). Tumor necrosis factor (TNF)-alpha and TNF receptors in viral pathogenesis. Proc. Soc. Exp. Biol. Med. 223, 241–257. doi: 10.1111/j.1525-1373.2000.22335.x
Hori, A., Kurata, S., and Kuraishi, T. (2018). Unexpected role of the IMD pathway in Drosophila gut defense against Staphylococcus aureus. Biochem. Biophys. Res. Commun. 495, 395–400. doi: 10.1016/j.bbrc.2017.11.004
Igboin, C. O., Moeschberger, M. L., Griffen, A. L., and Leys, E. J. (2011). Porphyromonas gingivalis virulence in a Drosophila melanogaster model. Infect. Immun. 79, 439–448. doi: 10.1128/IAI.00784-10
Jiang, H., Patel, P. H., Kohlmaier, A., Grenley, M. O., McEwen, D. G., and Edgar, B. A. (2009). Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343–1355. doi: 10.1016/j.cell.2009.05.014
Jones, T. A., Hernandez, D. Z., Wong, Z. C., Wandler, A. M., and Guillemin, K. (2017). The bacterial virulence factor CagA induces microbial dysbiosis that contributes to excessive epithelial cell proliferation in the Drosophila gut. PLoS Pathog. 13:e1006631. doi: 10.1371/journal.ppat.1006631
Kamareddine, L., Robins, W. P., Berkey, C. D., Mekalanos, J. J., and Watnick, P. I. (2018a). The Drosophila immune deficiency pathway modulates Enteroendocrine function and host metabolism. Cell Metab. 28, 449–462.e5. doi: 10.1016/j.cmet.2018.05.026
Kamareddine, L., Wong, A. C. N., Vanhove, A. S., Hang, S., Purdy, A. E., Kierek-Pearson, K., et al. (2018b). Activation of Vibrio cholerae quorum sensing promotes survival of an arthropod host. Nat. Microbiol. 3, 243–252. doi: 10.1038/s41564-017-0065-7
Kanoh, H., Kato, H., Suda, Y., Hori, A., Kurata, S., and Kuraishi, T. (2019). Dual comprehensive approach to decipher the Drosophila Toll pathway, ex vivo RNAi screenings and immunoprecipitation-mass spectrometry. Biochem. Biophys. Res. Commun. 508, 332–337. doi: 10.1016/j.bbrc.2018.11.007
Karlikow, M., Goic, B., and Saleh, M. C. (2014). RNAi and antiviral defense in Drosophila: setting up a systemic immune response. Dev. Comp. Immunol. 42, 85–92. doi: 10.1016/j.dci.2013.05.004
Kawai, T., and Akira, S. (2006). Innate immune recognition of viral infection. Nat. Immunol. 7, 131–137. doi: 10.1038/ni1303
Kemp, C., Mueller, S., Goto, A., Barbier, V., Paro, S., Bonnay, F., et al. (2013). Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J. Immunol. 190, 650–658. doi: 10.4049/jimmunol.1102486
Kiger, A. A., Baum, B., Jones, S., Jones, M. R., Coulson, A., Echeverri, C., et al. (2003). A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2:27. doi: 10.1186/1475-4924-2-27
Kim, S. H., Park, S. Y., Heo, Y. J., and Cho, Y. H. (2008). Drosophila melanogaster-based screening for multihost virulence factors of Pseudomonas aeruginosa PA14 and identification of a virulence-attenuating factor, HudA. Infect. Immun. 76, 4152–4162. doi: 10.1128/IAI.01637-07
Kocks, C., Cho, J. H., Nehme, N., Ulvila, J., Pearson, A. M., Meister, M., et al. (2005). Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell 123, 335–346. doi: 10.1016/j.cell.2005.08.034
Koo, I. C., Ohol, Y. M., Wu, P., Morisaki, J. H., Cox, J. S., and Brown, E. J. (2008). Role for lysosomal enzyme beta-hexosaminidase in the control of mycobacteria infection. Proc. Natl. Acad. Sci. U.S.A. 105, 710–715. doi: 10.1073/pnas.0708110105
Kounatidis, I., and Ligoxygakis, P. (2012). Drosophila as a model system to unravel the layers of innate immunity to infection. Open Biol. 2:120075. doi: 10.1098/rsob.120075
Kuttenkeuler, D., and Boutros, M. (2004). Genome-wide RNAi as a route to gene function in Drosophila. Brief. Funct. Genomic Proteomic 3, 168–176. doi: 10.1093/bfgp/3.2.168
Kuttenkeuler, D., Pelte, N., Ragab, A., Gesellchen, V., Schneider, L., Blass, C., et al. (2010). A large-scale RNAi screen identifies Deaf1 as a regulator of innate immune responses in Drosophila. J. Innate Immun. 2, 181–194. doi: 10.1159/000248649
Lacy, D. B., and Collier, R. J. (2002). Structure and function of anthrax toxin. Curr. Top. Microbiol. Immunol. 271, 61–85. doi: 10.1007/978-3-662-05767-4_4
Lamaris, G. A., Chamilos, G., Lewis, R. E., and Kontoyiannis, D. P. (2007). Virulence studies of Scedosporium and Fusarium species in Drosophila melanogaster. J. Infect. Dis. 196, 1860–1864. doi: 10.1086/523765
Leitão, A. B., Bian, X., Day, J. P., Pitton, S., Demir, E., and Jiggins, F. M. (2019). Independent effects on cellular and humoral immune responses underlie genotype-by-genotype interactions between Drosophila and parasitoids. PLoS Pathog. 15:e1008084. doi: 10.1371/journal.ppat.1008084
Lemaitre, B., and Hoffmann, J. (2007). The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743. doi: 10.1146/annurev.immunol.25.022106.141615
Leulier, F., Rodriguez, A., Khush, R. S., Abrams, J. M., and Lemaitre, B. (2000). The Drosophila caspase Dredd is required to resist gram-negative bacterial infection. EMBO Rep. 1, 353–358. doi: 10.1093/embo-reports/kvd073
Levashina, E. A., Moita, L. F., Blandin, S., Vriend, G., Lagueux, M., and Kafatos, F. C. (2001). Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles gambiae. Cell 104, 709–718. doi: 10.1016/S0092-8674(01)00267-7
Li, H., Li, W. X., and Ding, S. W. (2002). Induction and suppression of RNA silencing by an animal virus. Science 296, 1319–1321. doi: 10.1126/science.1070948
Liao, J.-F., Wu, C.-P., Tang, C.-K., Tsai, C.-W., Rouhová, L., and Wu, Y.-L. (2019). Identification of regulatory host genes involved in sigma virus replication using RNAi knockdown in Drosophila. Insects 10:339. doi: 10.3390/insects10100339
Ligoxygakis, P., Bulet, P., and Reichhart, J.-M. (2002a). Critical evaluation of the role of the Toll-like receptor 18-Wheeler in the host defense of Drosophila. EMBO Rep. 3, 666–673. doi: 10.1093/embo-reports/kvf130
Ligoxygakis, P., Pelte, N., Ji, C., Leclerc, V., Duvic, B., Belvin, M., et al. (2002b). A serpin mutant links Toll activation to melanization in the host defence of Drosophila. EMBO J. 21, 6330–6337. doi: 10.1093/emboj/cdf661
Lionakis, M. S., Lewis, R. E., May, G. S., Wiederhold, N. P., Albert, N. D., Halder, G., et al. (2005). Toll-deficient Drosophila flies as a fast, high-throughput model for the study of antifungal drug efficacy against invasive aspergillosis and Aspergillus virulence. J. Infect. Dis. 191, 1188–1195. doi: 10.1086/428587
Little, A. S., Okkotsu, Y., Reinhart, A. A., Damron, F. H., Barbier, M., Barrett, B., et al. (2018). Pseudomonas aeruginosa AlgR phosphorylation status differentially regulates pyocyanin and pyoverdine production. MBio 9:e02318-17. doi: 10.1128/mBio.02318-17
Lu, R., Maduro, M., Li, F., Li, H. W., Broitman-Maduro, G., Li, W. X., et al. (2005). Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature 436, 1040–1043. doi: 10.1038/nature03870
Martinez, E., and Campos-Gomez, J. (2016). Oxylipins produced by Pseudomonas aeruginosa promote biofilm formation and virulence. Nat. Commun. 7:13823. doi: 10.1038/ncomms13823
Medvedev, A. E., Murphy, M., Zhou, H., and Li, X. (2015). E3 ubiquitin ligases Pellinos as regulators of pattern recognition receptor signaling and immune responses. Immunol. Rev. 266, 109–122. doi: 10.1111/imr.12298
Medzhitov, R. (2001). Toll-like receptors and innate immunity. Nat. Rev. Immunol. 1, 135–145. doi: 10.1038/35100529
Meister, G., and Tuschl, T. (2004). Mechanisms of gene silencing by double-stranded RNA. Nature 431, 343–349. doi: 10.1038/nature02873
Meister, M., and Richards, G. (1996). Ecdysone and insect immunity: the maturation of the inducibility of the diptericin gene in Drosophila larvae. Insect Biochem. Mol. Biol. 26, 155–160. doi: 10.1016/0965-1748(95)00076-3
Merkel, S., Heidrich, D., Danilevicz, C. K., Scroferneker, M. L., and Zanette, R. A. (2018). Drosophila melanogaster as a model for the study of Malassezia pachydermatis infections. Vet. Microbiol. 224, 31–33. doi: 10.1016/j.vetmic.2018.08.021
Miyano-Kurosaki, N., and Takaku, H. (2006). “Gene silencing of virus replication by RNA interference,” in RNA Towards Medicine, eds V. Erdmann, J. Barciszewski, and J. Brosius (Poznan: Springer), 151–171. doi: 10.1007/3-540-27262-3_8
Myllymaki, H., and Ramet, M. (2014). JAK/STAT pathway in Drosophila immunity. Scand. J. Immunol. 79, 377–385. doi: 10.1111/sji.12170
Nappi, A. J., Carton, Y., and Frey, F. (1991). Parasite-induced enhancement of hemolymph tyrosinase activity in a selected immune reactive strain of Drosophila melanogaster. Archiv. Insect Biochem. Physiol. 18, 159–168. doi: 10.1002/arch.940180304
Nappi, A. J., and Vass, E. (1998). Hydrogen peroxide production in immune-reactive Drosophila melanogaster. J. Parasitol. 84, 1150–1157. doi: 10.2307/3284664
Nappi, A. J., Vass, E., Frey, F., and Carton, Y. (1995). Superoxide anion generation in Drosophila during melanotic encapsulation of parasites. Eur. J. Cell Biol. 68, 450–456.
Nappi, A. J., Vass, E., Frey, F., and Carton, Y. (2000). Nitric oxide involvement in Drosophila immunity. Nitric Oxide 4, 423–430. doi: 10.1006/niox.2000.0294
Nusslein-Volhard, C., and Wieschaus, E. (1980). Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801. doi: 10.1038/287795a0
Nykanen, A., Haley, B., and Zamore, P. D. (2001). ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107, 309–321. doi: 10.1016/S0092-8674(01)00547-5
Oh, C. T., Moon, C., Jeong, M. S., Kwon, S. H., and Jang, J. (2013). Drosophila melanogaster model for Mycobacterium abscessus infection. Microbes Infect. 15, 788–795. doi: 10.1016/j.micinf.2013.06.011
Paradkar, P. N., Trinidad, L., Voysey, R., Duchemin, J. B., and Walker, P. J. (2012). Secreted Vago restricts West Nile virus infection in Culex mosquito cells by activating the Jak-STAT pathway. Proc. Natl. Acad. Sci. U.S.A. 109, 18915–18920. doi: 10.1073/pnas.1205231109
Pearson, A., Lux, A., and Krieger, M. (1995). Expression cloning of dSR-CI, a class C macrophage-specific scavenger receptor from Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 92, 4056–4060. doi: 10.1073/pnas.92.9.4056
Pfeiffer, B. D., Ngo, T.-T. B., Hibbard, K. L., Murphy, C., Jenett, A., Truman, J. W., et al. (2010). Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735–755. doi: 10.1534/genetics.110.119917
Pham, L. N., Dionne, M. S., Shirasu-Hiza, M., and Schneider, D. S. (2007). A specific primed immune response in Drosophila is dependent on phagocytes. PLoS Pathog. 3:e26. doi: 10.1371/journal.ppat.0030026
Purdy, A. E., and Watnick, P. I. (2011). Spatially selective colonization of the arthropod intestine through activation of Vibrio cholerae biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 108, 19737–19742. doi: 10.1073/pnas.1111530108
Quintin, J., Asmar, J., Matskevich, A. A., Lafarge, M. C., and Ferrandon, D. (2013). The Drosophila Toll pathway controls but does not clear Candida glabrata infections. J. Immunol. 190, 2818–2827. doi: 10.4049/jimmunol.1201861
Ramet, M., Manfruelli, P., Pearson, A., Mathey-Prevot, B., and Ezekowitz, R. A. (2002). Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 416, 644–648. doi: 10.1038/nature735
Rizki, R. M., and Rizki, T. M. (1990). Parasitoid virus-like particles destroy Drosophila cellular immunity. Proc. Natl. Acad. Sci. U.S.A. 87, 8388–8392. doi: 10.1073/pnas.87.21.8388
Robles-Sikisaka, R., Garcia, D. K., Klimpel, K. R., and Dhar, A. K. (2001). Nucleotide sequence of 3′-end of the genome of Taura syndrome virus of shrimp suggests that it is related to insect picornaviruses. Arch. Virol. 146, 941–952. doi: 10.1007/s007050170126
Rose, P. P., Hanna, S. L., Spiridigliozzi, A., Wannissorn, N., Beiting, D. P., Ross, S. R., et al. (2011). Natural resistance-associated macrophage protein is a cellular receptor for sindbis virus in both insect and mammalian hosts. Cell Host Microbe 10, 97–104. doi: 10.1016/j.chom.2011.06.009
Rosetto, M., Engstrom, Y., Baldari, C. T., Telford, J. L., and Hultmark, D. (1995). Signals from the IL-1 receptor homolog, Toll, can activate an immune response in a Drosophila hemocyte cell line. Biochem. Biophys. Res. Commun. 209, 111–116. doi: 10.1006/bbrc.1995.1477
Sabin, L. R., Zhou, R., Gruber, J. J., Lukinova, N., Bambina, S., Berman, A., et al. (2009). Ars2 regulates both miRNA- and siRNA-dependent silencing and suppresses RNA virus infection in Drosophila. Cell 138, 340–351. doi: 10.1016/j.cell.2009.04.045
Saleh, M. C., Tassetto, M., van Rij, R. P., Goic, B., Gausson, V., Berry, B., et al. (2009). Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature 458, 346–350. doi: 10.1038/nature07712
Salt, G. (1970). The Cellular Defence Reactions of Insects. Vol. 16. Cambridge: Cambridge University Press.
Scherfer, C., Tang, H., Kambris, Z., Lhocine, N., Hashimoto, C., and Lemaitre, B. (2008). Drosophila Serpin-28D regulates hemolymph phenoloxidase activity and adult pigmentation. Dev. Biol. 323, 189–196. doi: 10.1016/j.ydbio.2008.08.030
Schmidt, R. L. (2014). A roadmap to understanding toll pathway changes: an educational primer for use with “regulation of toll signaling and inflammation by β-arrestin and the SUMO protease Ulp1”. Genetics 196, 923–929. doi: 10.1534/genetics.114.162289
Schmidt, R. L., Trejo, T. R., Plummer, T. B., Platt, J. L., and Tang, A. H. (2007). Infection-induced proteolysis of PGRP-LC controls the IMD activation and melanization cascades in Drosophila. FASEB J. 22, 918–929. doi: 10.1096/fj.06-7907com
Shelly, S., Lukinova, N., Bambina, S., Berman, A., and Cherry, S. (2009). Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity 30, 588–598. doi: 10.1016/j.immuni.2009.02.009
Shiratsuchi, A., Mori, T., Sakurai, K., Nagaosa, K., Sekimizu, K., Lee, B. L., et al. (2012). Independent recognition of Staphylococcus aureus by two receptors for phagocytosis in Drosophila. J. Biol. Chem. 287, 21663–21672. doi: 10.1074/jbc.M111.333807
Sjoberg, B. M., and Torrents, E. (2011). Shift in ribonucleotide reductase gene expression in Pseudomonas aeruginosa during infection. Infect. Immun. 79, 2663–2669. doi: 10.1128/IAI.01212-10
Söderhäll, K., and Cerenius, L. (1998). Role of the prophenoloxidase-activating system in invertebrate immunity. Curr. Opin. Immunol. 10, 23–28. doi: 10.1016/s0952-7915(98)80026-5
Song, M.-S., and Rossi, J. J. (2017). Molecular mechanisms of Dicer: endonuclease and enzymatic activity. Biochem. J. 474, 1603–1618. doi: 10.1042/BCJ20160759
Spikes, S., Xu, R., Nguyen, C. K., Chamilos, G., Kontoyiannis, D. P., Jacobson, R. H., et al. (2008). Gliotoxin production in Aspergillus fumigatus contributes to host-specific differences in virulence. J. Infect. Dis. 197, 479–486. doi: 10.1086/525044
Strand, M. R., and Pech, L. L. (1995). Immunological basis for compatibility in parasitoid-host relationships. Annu. Rev. Entomol. 40, 31–56. doi: 10.1146/annurev.en.40.010195.000335
Stroschein-Stevenson, S. L., Foley, E., O'Farrell, P. H., and Johnson, A. D. (2006). Identification of Drosophila gene products required for phagocytosis of Candida albicans. PLoS Biol. 4:e4. doi: 10.1371/journal.pbio.0040004
Tabuchi, Y., Shiratsuchi, A., Kurokawa, K., Gong, J. H., Sekimizu, K., Lee, B. L., et al. (2010). Inhibitory role for D-alanylation of wall teichoic acid in activation of insect Toll pathway by peptidoglycan of Staphylococcus aureus. J. Immunol. 185, 2424–2431. doi: 10.4049/jimmunol.1000625
Takehana, A., Katsuyama, T., Yano, T., Oshima, Y., Takada, H., Aigaki, T., et al. (2002). Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc. Natl. Acad. Sci. U.S.A. 99, 13705–13710. doi: 10.1073/pnas.212301199
Takehana, A., Yano, T., Mita, S., Kotani, A., Oshima, Y., and Kurata, S. (2004). Peptidoglycan recognition protein (PGRP)-LE and PGRP-LC act synergistically in Drosophila immunity. EMBO J. 23, 4690–4700. doi: 10.1038/sj.emboj.7600466
Tang, H., Kambris, Z., Lemaitre, B., and Hashimoto, C. (2008). A serpin that regulates immune melanization in the respiratory system of Drosophila. Dev. Cell 15, 617–626. doi: 10.1016/j.devcel.2008.08.017
Thevenon, D., Engel, E., Avet-Rochex, A., Gottar, M., Bergeret, E., Tricoire, H., et al. (2009). The Drosophila ubiquitin-specific protease dUSP36/Scny targets IMD to prevent constitutive immune signaling. Cell Host Microbe 6, 309–320. doi: 10.1016/j.chom.2009.09.007
Tsichritzis, T., Gaentzsch, P., Kosmidis, S., Brown, A., Skoulakis, E., Ligoxygakis, P., et al. (2007). A Drosophila ortholog of the human cylindromatosis tumor suppressor gene regulates triglyceride content and antibacterial defense. Development 134, 2605–2614. doi: 10.1242/dev.02859
Ueda, R. (2001). RNAi: a new technology in the post-genomic sequencing era. J. Neurogenet. 15, 193–204. doi: 10.3109/01677060109167376
van der Veen, A. G., Maillard, P. V., Schmidt, J. M., Lee, S. A., Deddouche-Grass, S., Borg, A., et al. (2018). The RIG-I-like receptor LGP2 inhibits Dicer-dependent processing of long double-stranded RNA and blocks RNA interference in mammalian cells. EMBO J. 37:e97479. doi: 10.15252/embj.201797479
Viswanatha, R., Brathwaite, R., Hu, Y., Li, Z., Rodiger, J., Merckaert, P., et al. (2019). Pooled CRISPR Screens in Drosophila cells. Curr. Protoc. Mol. Biol. 129:e111. doi: 10.1002/cpmb.111
Vodovar, N., Vinals, M., Liehl, P., Basset, A., Degrouard, J., Spellman, P., et al. (2005). Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc. Natl. Acad. Sci. U.S.A. 102, 11414–11419. doi: 10.1073/pnas.0502240102
Wang, X. H., Aliyari, R., Li, W. X., Li, H. W., Kim, K., Carthew, R., et al. (2006). RNA interference directs innate immunity against viruses in adult Drosophila. Science 312, 452–454. doi: 10.1126/science.1125694
Wang, Z., Hang, S., Purdy, A. E., and Watnick, P. I. (2013). Mutations in the IMD pathway and mustard counter Vibrio cholerae suppression of intestinal stem cell division in Drosophila. MBio 4:e00337-13. doi: 10.1128/mBio.00337-13
Waterfield, N., Sanchez-Contreras, M., Eleftherianos, I., Dowling, A., Yang, G., Wilkinson, P., et al. (2008). Rapid virulence annotation (RVA): identification of virulence factors using a bacterial genome library and multiple invertebrate hosts. Proc. Natl. Acad. Sci. U.S.A. 105, 15967–15972. doi: 10.1073/pnas.0711114105
Wilkins, C., Dishongh, R., Moore, S. C., Whitt, M. A., Chow, M., and Machaca, K. (2005). RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature 436, 1044–1047. doi: 10.1038/nature03957
Yagi, R., Mayer, F., and Basler, K. (2010). Refined LexA transactivators and their use in combination with the Drosophila Gal4 system. Proc. Natl. Acad. Sci. U.S.A. 107, 16166–16171. doi: 10.1073/pnas.1005957107
Yang, H., Kronhamn, J., Ekstrom, J. O., Korkut, G. G., and Hultmark, D. (2015). JAK/STAT signaling in Drosophila muscles controls the cellular immune response against parasitoid infection. EMBO Rep. 16, 1664–1672. doi: 10.15252/embr.201540277
Yassine, H., Kamareddine, L., and Osta, M. A. (2012). The mosquito melanization response is implicated in defense against the entomopathogenic fungus Beauveria bassiana. PLoS Pathog. 8:e1003029. doi: 10.1371/journal.ppat.1003029
Zambon, R. A., Nandakumar, M., Vakharia, V. N., and Wu, L. P. (2005). The Toll pathway is important for an antiviral response in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 102, 7257–7262. doi: 10.1073/pnas.0409181102
Zambon, R. A., Vakharia, V. N., and Wu, L. P. (2006). RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell Microbiol. 8, 880–889. doi: 10.1111/j.1462-5822.2006.00688.x
Keywords: Drosophila, host–pathogen interactions, host defense factors, pathogen virulence factors, disease control, disease progression
Citation: Younes S, Al-Sulaiti A, Nasser EAA, Najjar H and Kamareddine L (2020) Drosophila as a Model Organism in Host–Pathogen Interaction Studies. Front. Cell. Infect. Microbiol. 10:214. doi: 10.3389/fcimb.2020.00214
Received: 12 February 2020; Accepted: 20 April 2020;
Published: 23 June 2020.
Edited by:
Eric Ghigo, IHU Mediterranee Infection, FranceReviewed by:
Thomas Roeder, University of Kiel, GermanyShruti Yadav, Molecular Medicine Research Institute, United States
Copyright © 2020 Younes, Al-Sulaiti, Nasser, Najjar and Kamareddine. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Layla Kamareddine, lkamareddine@qu.edu.qa
†These authors have contributed equally to this work
 Salma Younes
Salma Younes Asma Al-Sulaiti
Asma Al-Sulaiti Elham Abdulwahab Ahmed Nasser†
Elham Abdulwahab Ahmed Nasser†  Hoda Najjar
Hoda Najjar Layla Kamareddine
Layla Kamareddine