Abstract
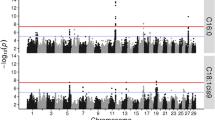
Milk fatty acid (FA) profile is a clear example of complex and multiple correlated traits whose genetic basis is difficult to assess. Although genome-wide association (GWA) studies have been successful in the identification of significant genetic variants for complex traits, when correlated phenotypes are analysed separately, the outcomes are difficult to compare and interpret in a metabolic context. Here, we performed a multivariate factor analysis (MFA) on Italian Simmental and Italian Holstein milk fat profiles to extract latent unobserved factors able to explain correlation structure and common metabolic function among different FAs. Individual factor scores obtained by MFA were used to perform a single-SNP based GWA. In both breeds, MFA was able to extract ten latent factors with specific biological meaning, notably: de novo synthesis, desaturation activity and biohydrogenation. The GWA result confirmed the increased power of joint association analysis on multiple correlated traits and allowed us to identify major candidate genes with well-documented function consistent with the metabolic classification of factors obtained, such as DGAT1, FASN and SCD.

Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19:1655–1664. https://doi.org/10.1101/gr.094052.109
Aulchenko YS, Ripke S, Isaacs A, van Duijn CM (2007) GenABEL: an R library for genome-wide association analysis. Bioinformatics 23:1294–1296. https://doi.org/10.1093/bioinformatics/btm108
Bauman DE, Mather IH, Wall RJ, Lock AL (2006) Major advances associated with the biosynthesis of milk. J Dairy Sci 89:1235–1243. https://doi.org/10.3168/jds.S0022-0302(06)72192-0
Bionaz M, Loor JJ (2008) Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genomics 9:366. https://doi.org/10.1186/1471-2164-9-366
Bouwman AC, Bovenhuis H, Visker MHPW, van Arendonk JAM (2011) Genome-wide association of milk fatty acids in Dutch dairy cattle. BMC Genet 12:43. https://doi.org/10.1186/1471-2156-12-43
Cecchinato A, Macciotta NPP, Mele M et al (2019) Genetic and genomic analyses of latent variables related to the milk fatty acid profile, milk composition, and udder health in dairy cattle. J Dairy Sci 102:5254–5265. https://doi.org/10.3168/jds.2018-15867
Cochran SD, Cole JB, Null DJ, Hansen PJ (2013) Discovery of single nucleotide polymorphisms in candidate genes associated with fertility and production traits in Holstein cattle. BMC Genet 14:49. https://doi.org/10.1186/1471-2156-14-49
Conte G, Dimauro C, Serra A et al (2018) A canonical discriminant analysis to study the association between milk fatty acids of ruminal origin and milk fat depression in dairy cows. J Dairy Sci 101:6497–6510. https://doi.org/10.3168/jds.2017-13941
Conte G, Serra A, Cremonesi P, et al (2016) Investigating mutual relationship among milk fatty acids by multivariate factor analysis in dairy cows. Livest Sci 188:124–132. /10.1016/j.livsci.2016.04.018
Fatumo S, Carstensen T, Nashiru O et al (2019) Complimentary methods for multivariate genome-wide association study identify new susceptibility genes for blood cell traits. Front Genet 10. https://doi.org/10.3389/fgene.2019.00334
Fievez V, Vlaeminck B, Dhanoa MS, Dewhurst RJ (2003) Use of principal component analysis to investigate the origin of heptadecenoic and conjugated linoleic acids in milk. J Dairy Sci 86:4047–4053. https://doi.org/10.3168/jds.S0022-0302(03)74016-8
Galesloot TE, van Steen K, Kiemeney LALM et al (2014) A comparison of multivariate genome-wide association methods. PLoS One 9:e95923. https://doi.org/10.1371/journal.pone.0095923
Gautier M, Barcelona RR, Fritz S et al (2006) Fine mapping and physical characterization of two linked quantitative trait loci affecting milk fat yield in dairy cattle on BTA26. Genetics 172:425–436. https://doi.org/10.1534/genetics.105.046169
Gómez-Cortés P, Juárez M, de la Fuente MA (2018) Milk fatty acids and potential health benefits: an updated vision. Trends Food Sci Tech 81:1–9. https://doi.org/10.1016/j.tifs.2018.08.014
Grisart B, Coppieters W, Farnir F et al (2002) Positional candidate cloning of a QTL in dairy cattle: identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res 12:222–231. https://doi.org/10.1101/gr.224202
Kaiser HF, Rice J (1974) Little jiffy, Mark Iv. Educ Psychol Meas 34:111–117. https://doi.org/10.1177/001316447403400115
Kaupe B, Winter A, Fries R, Erhardt G (2004) DGAT1 polymorphism in Bos indicus and Bos taurus cattle breeds. J Dairy Res 71:182–187. https://doi.org/10.1017/S0022029904000032
Kramer JKG, Hernandez M, Cruz-Hernandez C et al (2008) Combining results of two GC separations partly achieves determination of all cis and trans 16:1, 18:1, 18:2 and 18:3 except CLA isomers of milk fat as demonstrated using Ag-ion SPE fractionation. Lipids 43:259–273. https://doi.org/10.1007/s11745-007-3143-4
Macciotta NPP, Cecchinato A, Mele M, Bittante G (2012) Use of multivariate factor analysis to define new indicator variables for milk composition and coagulation properties in Brown Swiss cows. J Dairy Sci 95:7346–7354. https://doi.org/10.3168/jds.2012-5546
Macciotta NPP, Dimauro C, Null DJ, et al (2015) Dissection of genomic correlation matrices of US Holsteins using multivariate factor analysis. J Anim Breed Genet 132:9–20. https://doi.org/10.1111/jbg.12113
Mele M, Macciotta NPP, Cecchinato A et al (2016) Multivariate factor analysis of detailed milk fatty acid profile: effects of dairy system, feeding, herd, parity, and stage of lactation. J Dairy Sci 99:9820–9833. https://doi.org/10.3168/jds.2016-11451
Mele M, Serra A, Buccioni A et al (2008) Effect of soybean oil supplementation on milk fatty acid composition from Saanen goats fed diets with different forage:concentrate ratios. Ital J Anim Sci 7:297–311. https://doi.org/10.4081/ijas.2008.297
Morrison DF (1976) Multivariate statistical methods. McGraw-Hill Ryerson. /New York, NY
Nayeri S, Sargolzaei M, Abo-Ismail MK, et al (2017) Genome-wide association study for lactation persistency, female fertility, longevity, and lifetime profit index traits in Holstein dairy cattle. Journal of Dairy Science 100:1246–1258. https://doi.org/10.3168/jds.2016-11770
Nayeri S, Stothard P (2016) Tissues, metabolic pathways and genes of key importance in lactating dairy cattle. Springer Science Reviews 4:49–77. https://doi.org/10.1007/s40362-016-0040-3
Palombo V, Milanesi M, Sgorlon S, et al (2018) Genome-wide association study of milk fatty acid composition in Italian Simmental and Italian Holstein cows using single nucleotide polymorphism arrays. J Dairy Sci. https://doi.org/10.3168/jds.2018-14413
Pulina G, Francesconi AHD, Stefanon B, et al (2017) Sustainable ruminant production to help feed the planet. Italian Journal of Animal Science 16:140–171. https://doi.org/10.1080/1828051X.2016.1260500
R Core Team (2018) R: a language and environment for statistical computing. R Foundation for Statistical Computing Vienna, Austria https://www.R-project.org/
Raven L-A, Cocks BG, Hayes BJ (2014) Multibreed genome wide association can improve precision of mapping causative variants underlying milk production in dairy cattle. BMC Genomics 15:62. https://doi.org/10.1186/1471-2164-15-62
Scotti E, Fontanesi L, Schiavini F, et al (2010) DGAT1 p.K232A polymorphism in dairy and dual purpose Italian cattle breeds. Ital J Anim Sci 9:e16. https://doi.org/10.4081/10.4081/ijas.2010.e16
Serra A, Conte G, Ciucci F, et al (2018) Dietary linseed supplementation affects the fatty acid composition of the sn-2 position of triglycerides in sheep milk. J Dairy Sci 101:6742–6751. /https://doi.org/10.3168/jds.2017-14188
Shingfield KJ, Bernard L, Leroux C, Chilliard Y (2010) Role of trans fatty acids in the nutritional regulation of mammary lipogenesis in ruminants. Animal 4:1140–1166. https://doi.org/10.1017/S1751731110000510
Shingfield KJ, Bonnet M, Scollan ND (2013) Recent developments in altering the fatty acid composition of ruminant-derived foods. Animal 7 Suppl 1:132–162. https://doi.org/10.1017/S1751731112001681
Soyeurt H, Gillon A, Vanderick S, et al (2007) Estimation of heritability and genetic correlations for the major fatty acids in bovine milk. Journal of Dairy Science 90:4435–4442. https://doi.org/10.3168/jds.2007-0054
Suburu J, Shi L, Wu J et al (2014) Fatty acid synthase is required for mammary gland development and milk production during lactation. Am J Physiol Endocrinol Metab 306:E1132–E1143. https://doi.org/10.1152/ajpendo.00514.2013
Vlaeminck B, Fievez V, Tamminga S, et al (2006) Milk odd- and branched-chain fatty acids in relation to the rumen fermentation pattern. J Dairy Sci 89:3954–3964. https://doi.org/10.3168/jds.S0022-0302(06)72437-7
Wang X, Wurmser C, Pausch H et al (2012) Identification and dissection of four major QTL affecting milk fat content in the German Holstein-Friesian population. PLoS One 7:e40711. https://doi.org/10.1371/journal.pone.0040711
Funding
Research was supported by the Italian Ministero dell’Istruzione, dell’Università e della Ricerca—MIUR (Rome, Italy; PRIN—GEN2PHEN project).
Author information
Authors and Affiliations
Contributions
MD, PAM and NPPM conceived and designed the experiment. MD, MM and BS performed the experiment. VP and GC analysed the data. MD and MM interpreted the results. VP and GC wrote the paper. MD, BS, NPPM and PAM edited and reviewed the manuscript. All the authors read and approved the final manuscript. VP and GC contributed equally to this paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Ethics approval
All farms adhere to a high standard of veterinary care based on best practice manual under the supervision of the official veterinary service and according to European directives and laws. Sample collection was approved by the Bioethics Committee of the University of Udine.
Consent for publication
Not applicable.
Additional information
Communicated by: Maciej Szydlowski
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
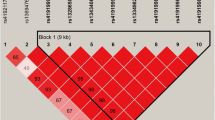
Correlation and partial correlation heatmap for the 42 fatty acids used in the multivariate factor analysis in Italian Simmental breed. Upper diagonal elements are the Pearson correlation coefficients; lower diagonal elements are the partial correlation coefficients. Positive correlations are displayed in blue and negative correlations in red color. Color intensity is proportional to the correlation value. (DOCX 89 kb)
ESM 2
Correlation and partial correlation heatmap for the 42 fatty acids used in the multivariate factor analysis in Italian Holstein breed. Upper diagonal elements are the Pearson correlation coefficients; lower diagonal elements are the partial correlation coefficients. Positive correlations are displayed in blue and negative correlations in red color. Color intensity is proportional to the correlation value. (DOCX 89 kb)
ESM 3
Correlation and partial correlation matrix for the 42 fatty acids used in the multivariate factor analysis in Italian Simmental breed. Upper diagonal elements are the Pearson correlation coefficients; lower diagonal elements are the partial correlation coefficients. (DOCX 76 kb)
ESM 4
Correlation and partial correlation matrix for the 42 fatty acids used in the multivariate factor analysis in Italian Holstein breed. Upper diagonal elements are the Pearson correlation coefficients; lower diagonal elements are the partial correlation coefficients. (XLSX 36 kb)
ESM 5
List of genes pinpointed considering boundaries of < 100 kbp (kilo base pairs) on both sides from significant SNPs (Bos taurus ARS-UCD1.2 genome assembly). (XLSX 36 kb)
Rights and permissions
About this article
Cite this article
Palombo, V., Conte, G., Mele, M. et al. Use of multivariate factor analysis of detailed milk fatty acid profile to perform a genome-wide association study in Italian Simmental and Italian Holstein. J Appl Genetics 61, 451–463 (2020). https://doi.org/10.1007/s13353-020-00568-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-020-00568-2