Abstract
Objectives
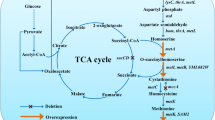
To improve the S-adenosylmethionine (SAM) production in methionine-free medium, effects of deleting genes of SAM decarboxylase (speD) and homoserine kinase (thrB) on SAM titers were investigated, and the SAM synthetase gene (SAM2) was also overexpressed.
Results
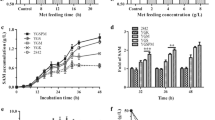
In B. amyloliquefaciens HSAM2, deleting speD to block the SAM utilization pathway significantly reduced the SAM titer. After knockout of thrB to block the branched pathway, the resulted mutant HSAM4 produced 143.93 mg/L SAM, increasing by 42% than HSAM2. Further plasmid-based expression of SAM2 improved the SAM titer to 226.92 mg/L, and final optimization of key fermentation parameters resulted in the maximum SAM titer of 412.01 mg/L in flasks batch fermentation.
Conclusions
Deleting thrB and overexpressing SAM2 gene were efficient for enhanced SAM production in B. amyloliquefaciens. The maximum SAM titer in flasks batch fermentation was much higher than that of previous reports.





Similar content being viewed by others
References
Cederbaum AI (2010) Hepatoprotective effects of S-adenosyl-L-methionine against alcohol- and cytochrome P450 2E1-induced liver injury. World J Gastroenterol 16:1366–1376
Chen Y, Lou S, Fan L, Zhang X, Tan T (2015a) Control of ATP concentration in Escherichia coli using synthetic small regulatory RNAs for enhanced S-adenosylmethionine production. FEMS Microbiol Lett 362:fnv115
Chen Y, Xu D, Fan L, Zhang X, Tan T (2015b) Manipulating multi-system of NADPH regulation in Escherichia coli for enhanced S-adenosylmethionine production. RSC Adv 5:41103–41111
Chu J, Qian J, Zhuang Y, Zhang S, Li Y (2013) Progress in the research of S-adenosyl-L-methionine production. Appl Microbiol Biotechnol 97:41–49
Fontecave M, Atta M, Mulliez E (2004) S-adenosylmethionine: nothing goes to waste. Trends Biochem Sci 29:243–249
Han G, Hu X, Qin T, Li Y, Wang X (2016) Metabolic engineering of Corynebacterium glutamicum ATCC13032 to produce S-adenosyl-L-methionine. Enzyme Microb Technol 83:14–21
Kanai M, Mizunuma M, Fujii T, Iefuji H (2017) A genetic method to enhance the accumulation of S-adenosylmethionine in yeast. Appl Microbiol Biotechnol 101:1351–1357
Li B, Kurihara S, Kim SH, Liang J, Michael AJ (2019) A polyamine-independent role for S-adenosylmethionine decarboxylase. Biochem J 476:2579–2594
Ren W, Cai D, Hu S, Xia S, Wang Z, Tan T, Zhang Q (2017) S-Adenosyl-L-methionine production by Saccharomyces cerevisiae SAM 0801 using DL-methionine mixture: from laboratory to pilot scale. Process Biochem 62:48–52
Ruan L et al (2019) Metabolic engineering of Bacillus amyloliquefaciens for enhanced production of S-adenosylmethionine by coupling of an engineered S-adenosylmethionine pathway and the tricarboxylic acid cycle. Biotechnol Biofuels 12:211
Walther T et al (2017) Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor 2,4-dihydroxybutyric acid. Nat Commun 8:1–12
Williams AL, Girard C, Jui D, Sabina A, Katz DL (2005) S-Adenosylmethionine (SAMe) as treatment for depression: a systematic review. Clin Invest Med 28:132–139
Zhao W, Hang B, Zhu X, Wang R, Shen M, Huang L, Xu Z (2016) Improving the productivity of S-adenosyl-L-methionine by metabolic engineering in an industrial Saccharomyces cerevisiae strain. J Biotechnol 236:64–70
Supporting information
Supplementary Table 1—Primers used for PCR in this study.
Funding
This work was supported by Da Bei Nong Group Promoted Project for Young Scholar of HZAU (No. 2017DBN011) and the National Natural Science Foundation of China (31501468).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jiang, C., Ruan, L., Wei, X. et al. Enhancement of S-adenosylmethionine production by deleting thrB gene and overexpressing SAM2 gene in Bacillus amyloliquefaciens. Biotechnol Lett 42, 2293–2298 (2020). https://doi.org/10.1007/s10529-020-02945-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-020-02945-7